Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
- Dissertation
- How to Write a Results Section | Tips & Examples

How to Write a Results Section | Tips & Examples
Published on August 30, 2022 by Tegan George . Revised on July 18, 2023.
A results section is where you report the main findings of the data collection and analysis you conducted for your thesis or dissertation . You should report all relevant results concisely and objectively, in a logical order. Don’t include subjective interpretations of why you found these results or what they mean—any evaluation should be saved for the discussion section .
Instantly correct all language mistakes in your text
Upload your document to correct all your mistakes in minutes
Table of contents
How to write a results section, reporting quantitative research results, reporting qualitative research results, results vs. discussion vs. conclusion, checklist: research results, other interesting articles, frequently asked questions about results sections.
When conducting research, it’s important to report the results of your study prior to discussing your interpretations of it. This gives your reader a clear idea of exactly what you found and keeps the data itself separate from your subjective analysis.
Here are a few best practices:
- Your results should always be written in the past tense.
- While the length of this section depends on how much data you collected and analyzed, it should be written as concisely as possible.
- Only include results that are directly relevant to answering your research questions . Avoid speculative or interpretative words like “appears” or “implies.”
- If you have other results you’d like to include, consider adding them to an appendix or footnotes.
- Always start out with your broadest results first, and then flow into your more granular (but still relevant) ones. Think of it like a shoe store: first discuss the shoes as a whole, then the sneakers, boots, sandals, etc.
Here's why students love Scribbr's proofreading services
Discover proofreading & editing
If you conducted quantitative research , you’ll likely be working with the results of some sort of statistical analysis .
Your results section should report the results of any statistical tests you used to compare groups or assess relationships between variables . It should also state whether or not each hypothesis was supported.
The most logical way to structure quantitative results is to frame them around your research questions or hypotheses. For each question or hypothesis, share:
- A reminder of the type of analysis you used (e.g., a two-sample t test or simple linear regression ). A more detailed description of your analysis should go in your methodology section.
- A concise summary of each relevant result, both positive and negative. This can include any relevant descriptive statistics (e.g., means and standard deviations ) as well as inferential statistics (e.g., t scores, degrees of freedom , and p values ). Remember, these numbers are often placed in parentheses.
- A brief statement of how each result relates to the question, or whether the hypothesis was supported. You can briefly mention any results that didn’t fit with your expectations and assumptions, but save any speculation on their meaning or consequences for your discussion and conclusion.
A note on tables and figures
In quantitative research, it’s often helpful to include visual elements such as graphs, charts, and tables , but only if they are directly relevant to your results. Give these elements clear, descriptive titles and labels so that your reader can easily understand what is being shown. If you want to include any other visual elements that are more tangential in nature, consider adding a figure and table list .
As a rule of thumb:
- Tables are used to communicate exact values, giving a concise overview of various results
- Graphs and charts are used to visualize trends and relationships, giving an at-a-glance illustration of key findings
Don’t forget to also mention any tables and figures you used within the text of your results section. Summarize or elaborate on specific aspects you think your reader should know about rather than merely restating the same numbers already shown.
A two-sample t test was used to test the hypothesis that higher social distance from environmental problems would reduce the intent to donate to environmental organizations, with donation intention (recorded as a score from 1 to 10) as the outcome variable and social distance (categorized as either a low or high level of social distance) as the predictor variable.Social distance was found to be positively correlated with donation intention, t (98) = 12.19, p < .001, with the donation intention of the high social distance group 0.28 points higher, on average, than the low social distance group (see figure 1). This contradicts the initial hypothesis that social distance would decrease donation intention, and in fact suggests a small effect in the opposite direction.
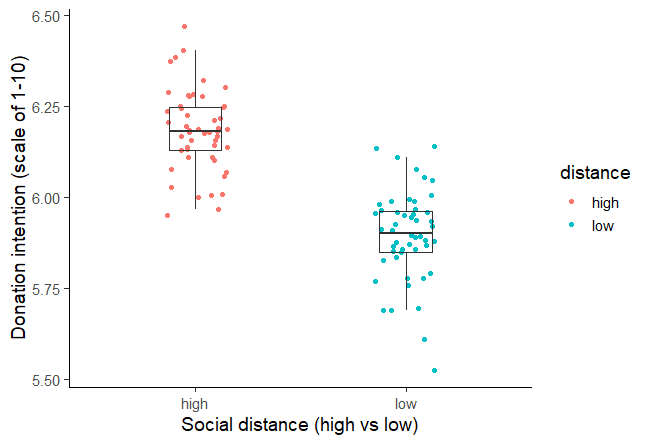
Figure 1: Intention to donate to environmental organizations based on social distance from impact of environmental damage.
In qualitative research , your results might not all be directly related to specific hypotheses. In this case, you can structure your results section around key themes or topics that emerged from your analysis of the data.
For each theme, start with general observations about what the data showed. You can mention:
- Recurring points of agreement or disagreement
- Patterns and trends
- Particularly significant snippets from individual responses
Next, clarify and support these points with direct quotations. Be sure to report any relevant demographic information about participants. Further information (such as full transcripts , if appropriate) can be included in an appendix .
When asked about video games as a form of art, the respondents tended to believe that video games themselves are not an art form, but agreed that creativity is involved in their production. The criteria used to identify artistic video games included design, story, music, and creative teams.One respondent (male, 24) noted a difference in creativity between popular video game genres:
“I think that in role-playing games, there’s more attention to character design, to world design, because the whole story is important and more attention is paid to certain game elements […] so that perhaps you do need bigger teams of creative experts than in an average shooter or something.”
Responses suggest that video game consumers consider some types of games to have more artistic potential than others.
Your results section should objectively report your findings, presenting only brief observations in relation to each question, hypothesis, or theme.
It should not speculate about the meaning of the results or attempt to answer your main research question . Detailed interpretation of your results is more suitable for your discussion section , while synthesis of your results into an overall answer to your main research question is best left for your conclusion .
Receive feedback on language, structure, and formatting
Professional editors proofread and edit your paper by focusing on:
- Academic style
- Vague sentences
- Style consistency
See an example
I have completed my data collection and analyzed the results.
I have included all results that are relevant to my research questions.
I have concisely and objectively reported each result, including relevant descriptive statistics and inferential statistics .
I have stated whether each hypothesis was supported or refuted.
I have used tables and figures to illustrate my results where appropriate.
All tables and figures are correctly labelled and referred to in the text.
There is no subjective interpretation or speculation on the meaning of the results.
You've finished writing up your results! Use the other checklists to further improve your thesis.
If you want to know more about AI for academic writing, AI tools, or research bias, make sure to check out some of our other articles with explanations and examples or go directly to our tools!
Research bias
- Survivorship bias
- Self-serving bias
- Availability heuristic
- Halo effect
- Hindsight bias
- Deep learning
- Generative AI
- Machine learning
- Reinforcement learning
- Supervised vs. unsupervised learning
(AI) Tools
- Grammar Checker
- Paraphrasing Tool
- Text Summarizer
- AI Detector
- Plagiarism Checker
- Citation Generator
The results chapter of a thesis or dissertation presents your research results concisely and objectively.
In quantitative research , for each question or hypothesis , state:
- The type of analysis used
- Relevant results in the form of descriptive and inferential statistics
- Whether or not the alternative hypothesis was supported
In qualitative research , for each question or theme, describe:
- Recurring patterns
- Significant or representative individual responses
- Relevant quotations from the data
Don’t interpret or speculate in the results chapter.
Results are usually written in the past tense , because they are describing the outcome of completed actions.
The results chapter or section simply and objectively reports what you found, without speculating on why you found these results. The discussion interprets the meaning of the results, puts them in context, and explains why they matter.
In qualitative research , results and discussion are sometimes combined. But in quantitative research , it’s considered important to separate the objective results from your interpretation of them.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
George, T. (2023, July 18). How to Write a Results Section | Tips & Examples. Scribbr. Retrieved April 9, 2024, from https://www.scribbr.com/dissertation/results/
Is this article helpful?
Tegan George
Other students also liked, what is a research methodology | steps & tips, how to write a discussion section | tips & examples, how to write a thesis or dissertation conclusion, "i thought ai proofreading was useless but..".
I've been using Scribbr for years now and I know it's a service that won't disappoint. It does a good job spotting mistakes”
Editorial Manager, our manuscript submissions site will be unavailable between 12pm April 5, 2024 and 12pm April 8 2024 (Pacific Standard Time). We apologize for any inconvenience this may cause.
When you choose to publish with PLOS, your research makes an impact. Make your work accessible to all, without restrictions, and accelerate scientific discovery with options like preprints and published peer review that make your work more Open.
- PLOS Biology
- PLOS Climate
- PLOS Complex Systems
- PLOS Computational Biology
- PLOS Digital Health
- PLOS Genetics
- PLOS Global Public Health
- PLOS Medicine
- PLOS Mental Health
- PLOS Neglected Tropical Diseases
- PLOS Pathogens
- PLOS Sustainability and Transformation
- PLOS Collections
- How to Write Discussions and Conclusions

The discussion section contains the results and outcomes of a study. An effective discussion informs readers what can be learned from your experiment and provides context for the results.
What makes an effective discussion?
When you’re ready to write your discussion, you’ve already introduced the purpose of your study and provided an in-depth description of the methodology. The discussion informs readers about the larger implications of your study based on the results. Highlighting these implications while not overstating the findings can be challenging, especially when you’re submitting to a journal that selects articles based on novelty or potential impact. Regardless of what journal you are submitting to, the discussion section always serves the same purpose: concluding what your study results actually mean.
A successful discussion section puts your findings in context. It should include:
- the results of your research,
- a discussion of related research, and
- a comparison between your results and initial hypothesis.
Tip: Not all journals share the same naming conventions.
You can apply the advice in this article to the conclusion, results or discussion sections of your manuscript.
Our Early Career Researcher community tells us that the conclusion is often considered the most difficult aspect of a manuscript to write. To help, this guide provides questions to ask yourself, a basic structure to model your discussion off of and examples from published manuscripts.
Questions to ask yourself:
- Was my hypothesis correct?
- If my hypothesis is partially correct or entirely different, what can be learned from the results?
- How do the conclusions reshape or add onto the existing knowledge in the field? What does previous research say about the topic?
- Why are the results important or relevant to your audience? Do they add further evidence to a scientific consensus or disprove prior studies?
- How can future research build on these observations? What are the key experiments that must be done?
- What is the “take-home” message you want your reader to leave with?
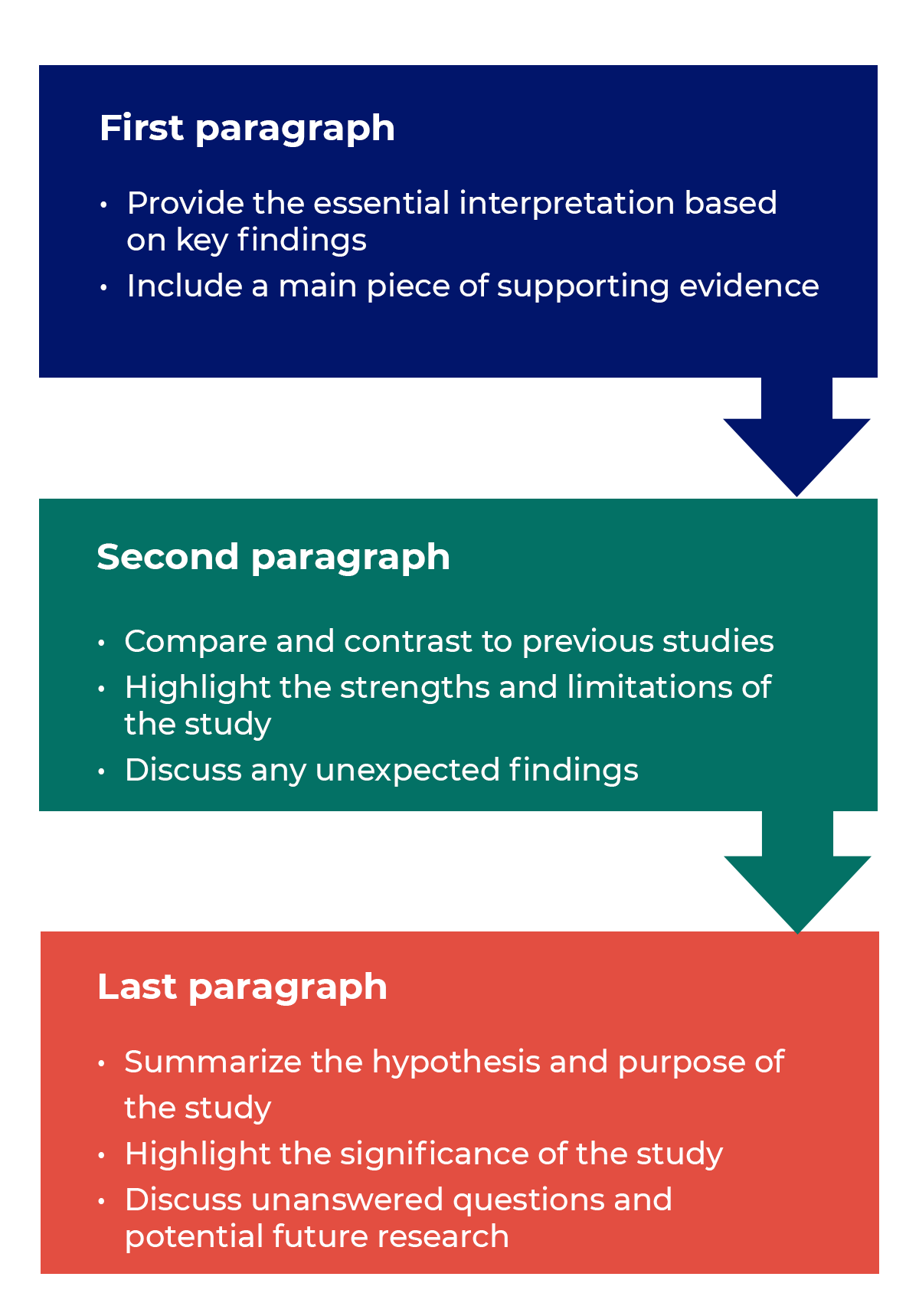
How to structure a discussion
Trying to fit a complete discussion into a single paragraph can add unnecessary stress to the writing process. If possible, you’ll want to give yourself two or three paragraphs to give the reader a comprehensive understanding of your study as a whole. Here’s one way to structure an effective discussion:
Writing Tips
While the above sections can help you brainstorm and structure your discussion, there are many common mistakes that writers revert to when having difficulties with their paper. Writing a discussion can be a delicate balance between summarizing your results, providing proper context for your research and avoiding introducing new information. Remember that your paper should be both confident and honest about the results!

- Read the journal’s guidelines on the discussion and conclusion sections. If possible, learn about the guidelines before writing the discussion to ensure you’re writing to meet their expectations.
- Begin with a clear statement of the principal findings. This will reinforce the main take-away for the reader and set up the rest of the discussion.
- Explain why the outcomes of your study are important to the reader. Discuss the implications of your findings realistically based on previous literature, highlighting both the strengths and limitations of the research.
- State whether the results prove or disprove your hypothesis. If your hypothesis was disproved, what might be the reasons?
- Introduce new or expanded ways to think about the research question. Indicate what next steps can be taken to further pursue any unresolved questions.
- If dealing with a contemporary or ongoing problem, such as climate change, discuss possible consequences if the problem is avoided.
- Be concise. Adding unnecessary detail can distract from the main findings.

Don’t
- Rewrite your abstract. Statements with “we investigated” or “we studied” generally do not belong in the discussion.
- Include new arguments or evidence not previously discussed. Necessary information and evidence should be introduced in the main body of the paper.
- Apologize. Even if your research contains significant limitations, don’t undermine your authority by including statements that doubt your methodology or execution.
- Shy away from speaking on limitations or negative results. Including limitations and negative results will give readers a complete understanding of the presented research. Potential limitations include sources of potential bias, threats to internal or external validity, barriers to implementing an intervention and other issues inherent to the study design.
- Overstate the importance of your findings. Making grand statements about how a study will fully resolve large questions can lead readers to doubt the success of the research.
Snippets of Effective Discussions:
Consumer-based actions to reduce plastic pollution in rivers: A multi-criteria decision analysis approach
Identifying reliable indicators of fitness in polar bears
- How to Write a Great Title
- How to Write an Abstract
- How to Write Your Methods
- How to Report Statistics
- How to Edit Your Work
The contents of the Peer Review Center are also available as a live, interactive training session, complete with slides, talking points, and activities. …
The contents of the Writing Center are also available as a live, interactive training session, complete with slides, talking points, and activities. …
There’s a lot to consider when deciding where to submit your work. Learn how to choose a journal that will help your study reach its audience, while reflecting your values as a researcher…
- Privacy Policy
Buy Me a Coffee

Home » Research Results Section – Writing Guide and Examples
Research Results Section – Writing Guide and Examples
Table of Contents

Research Results
Research results refer to the findings and conclusions derived from a systematic investigation or study conducted to answer a specific question or hypothesis. These results are typically presented in a written report or paper and can include various forms of data such as numerical data, qualitative data, statistics, charts, graphs, and visual aids.
Results Section in Research
The results section of the research paper presents the findings of the study. It is the part of the paper where the researcher reports the data collected during the study and analyzes it to draw conclusions.
In the results section, the researcher should describe the data that was collected, the statistical analysis performed, and the findings of the study. It is important to be objective and not interpret the data in this section. Instead, the researcher should report the data as accurately and objectively as possible.
Structure of Research Results Section
The structure of the research results section can vary depending on the type of research conducted, but in general, it should contain the following components:
- Introduction: The introduction should provide an overview of the study, its aims, and its research questions. It should also briefly explain the methodology used to conduct the study.
- Data presentation : This section presents the data collected during the study. It may include tables, graphs, or other visual aids to help readers better understand the data. The data presented should be organized in a logical and coherent way, with headings and subheadings used to help guide the reader.
- Data analysis: In this section, the data presented in the previous section are analyzed and interpreted. The statistical tests used to analyze the data should be clearly explained, and the results of the tests should be presented in a way that is easy to understand.
- Discussion of results : This section should provide an interpretation of the results of the study, including a discussion of any unexpected findings. The discussion should also address the study’s research questions and explain how the results contribute to the field of study.
- Limitations: This section should acknowledge any limitations of the study, such as sample size, data collection methods, or other factors that may have influenced the results.
- Conclusions: The conclusions should summarize the main findings of the study and provide a final interpretation of the results. The conclusions should also address the study’s research questions and explain how the results contribute to the field of study.
- Recommendations : This section may provide recommendations for future research based on the study’s findings. It may also suggest practical applications for the study’s results in real-world settings.
Outline of Research Results Section
The following is an outline of the key components typically included in the Results section:
I. Introduction
- A brief overview of the research objectives and hypotheses
- A statement of the research question
II. Descriptive statistics
- Summary statistics (e.g., mean, standard deviation) for each variable analyzed
- Frequencies and percentages for categorical variables
III. Inferential statistics
- Results of statistical analyses, including tests of hypotheses
- Tables or figures to display statistical results
IV. Effect sizes and confidence intervals
- Effect sizes (e.g., Cohen’s d, odds ratio) to quantify the strength of the relationship between variables
- Confidence intervals to estimate the range of plausible values for the effect size
V. Subgroup analyses
- Results of analyses that examined differences between subgroups (e.g., by gender, age, treatment group)
VI. Limitations and assumptions
- Discussion of any limitations of the study and potential sources of bias
- Assumptions made in the statistical analyses
VII. Conclusions
- A summary of the key findings and their implications
- A statement of whether the hypotheses were supported or not
- Suggestions for future research
Example of Research Results Section
An Example of a Research Results Section could be:
- This study sought to examine the relationship between sleep quality and academic performance in college students.
- Hypothesis : College students who report better sleep quality will have higher GPAs than those who report poor sleep quality.
- Methodology : Participants completed a survey about their sleep habits and academic performance.
II. Participants
- Participants were college students (N=200) from a mid-sized public university in the United States.
- The sample was evenly split by gender (50% female, 50% male) and predominantly white (85%).
- Participants were recruited through flyers and online advertisements.
III. Results
- Participants who reported better sleep quality had significantly higher GPAs (M=3.5, SD=0.5) than those who reported poor sleep quality (M=2.9, SD=0.6).
- See Table 1 for a summary of the results.
- Participants who reported consistent sleep schedules had higher GPAs than those with irregular sleep schedules.
IV. Discussion
- The results support the hypothesis that better sleep quality is associated with higher academic performance in college students.
- These findings have implications for college students, as prioritizing sleep could lead to better academic outcomes.
- Limitations of the study include self-reported data and the lack of control for other variables that could impact academic performance.
V. Conclusion
- College students who prioritize sleep may see a positive impact on their academic performance.
- These findings highlight the importance of sleep in academic success.
- Future research could explore interventions to improve sleep quality in college students.
Example of Research Results in Research Paper :
Our study aimed to compare the performance of three different machine learning algorithms (Random Forest, Support Vector Machine, and Neural Network) in predicting customer churn in a telecommunications company. We collected a dataset of 10,000 customer records, with 20 predictor variables and a binary churn outcome variable.
Our analysis revealed that all three algorithms performed well in predicting customer churn, with an overall accuracy of 85%. However, the Random Forest algorithm showed the highest accuracy (88%), followed by the Support Vector Machine (86%) and the Neural Network (84%).
Furthermore, we found that the most important predictor variables for customer churn were monthly charges, contract type, and tenure. Random Forest identified monthly charges as the most important variable, while Support Vector Machine and Neural Network identified contract type as the most important.
Overall, our results suggest that machine learning algorithms can be effective in predicting customer churn in a telecommunications company, and that Random Forest is the most accurate algorithm for this task.
Example 3 :
Title : The Impact of Social Media on Body Image and Self-Esteem
Abstract : This study aimed to investigate the relationship between social media use, body image, and self-esteem among young adults. A total of 200 participants were recruited from a university and completed self-report measures of social media use, body image satisfaction, and self-esteem.
Results: The results showed that social media use was significantly associated with body image dissatisfaction and lower self-esteem. Specifically, participants who reported spending more time on social media platforms had lower levels of body image satisfaction and self-esteem compared to those who reported less social media use. Moreover, the study found that comparing oneself to others on social media was a significant predictor of body image dissatisfaction and lower self-esteem.
Conclusion : These results suggest that social media use can have negative effects on body image satisfaction and self-esteem among young adults. It is important for individuals to be mindful of their social media use and to recognize the potential negative impact it can have on their mental health. Furthermore, interventions aimed at promoting positive body image and self-esteem should take into account the role of social media in shaping these attitudes and behaviors.
Importance of Research Results
Research results are important for several reasons, including:
- Advancing knowledge: Research results can contribute to the advancement of knowledge in a particular field, whether it be in science, technology, medicine, social sciences, or humanities.
- Developing theories: Research results can help to develop or modify existing theories and create new ones.
- Improving practices: Research results can inform and improve practices in various fields, such as education, healthcare, business, and public policy.
- Identifying problems and solutions: Research results can identify problems and provide solutions to complex issues in society, including issues related to health, environment, social justice, and economics.
- Validating claims : Research results can validate or refute claims made by individuals or groups in society, such as politicians, corporations, or activists.
- Providing evidence: Research results can provide evidence to support decision-making, policy-making, and resource allocation in various fields.
How to Write Results in A Research Paper
Here are some general guidelines on how to write results in a research paper:
- Organize the results section: Start by organizing the results section in a logical and coherent manner. Divide the section into subsections if necessary, based on the research questions or hypotheses.
- Present the findings: Present the findings in a clear and concise manner. Use tables, graphs, and figures to illustrate the data and make the presentation more engaging.
- Describe the data: Describe the data in detail, including the sample size, response rate, and any missing data. Provide relevant descriptive statistics such as means, standard deviations, and ranges.
- Interpret the findings: Interpret the findings in light of the research questions or hypotheses. Discuss the implications of the findings and the extent to which they support or contradict existing theories or previous research.
- Discuss the limitations : Discuss the limitations of the study, including any potential sources of bias or confounding factors that may have affected the results.
- Compare the results : Compare the results with those of previous studies or theoretical predictions. Discuss any similarities, differences, or inconsistencies.
- Avoid redundancy: Avoid repeating information that has already been presented in the introduction or methods sections. Instead, focus on presenting new and relevant information.
- Be objective: Be objective in presenting the results, avoiding any personal biases or interpretations.
When to Write Research Results
Here are situations When to Write Research Results”
- After conducting research on the chosen topic and obtaining relevant data, organize the findings in a structured format that accurately represents the information gathered.
- Once the data has been analyzed and interpreted, and conclusions have been drawn, begin the writing process.
- Before starting to write, ensure that the research results adhere to the guidelines and requirements of the intended audience, such as a scientific journal or academic conference.
- Begin by writing an abstract that briefly summarizes the research question, methodology, findings, and conclusions.
- Follow the abstract with an introduction that provides context for the research, explains its significance, and outlines the research question and objectives.
- The next section should be a literature review that provides an overview of existing research on the topic and highlights the gaps in knowledge that the current research seeks to address.
- The methodology section should provide a detailed explanation of the research design, including the sample size, data collection methods, and analytical techniques used.
- Present the research results in a clear and concise manner, using graphs, tables, and figures to illustrate the findings.
- Discuss the implications of the research results, including how they contribute to the existing body of knowledge on the topic and what further research is needed.
- Conclude the paper by summarizing the main findings, reiterating the significance of the research, and offering suggestions for future research.
Purpose of Research Results
The purposes of Research Results are as follows:
- Informing policy and practice: Research results can provide evidence-based information to inform policy decisions, such as in the fields of healthcare, education, and environmental regulation. They can also inform best practices in fields such as business, engineering, and social work.
- Addressing societal problems : Research results can be used to help address societal problems, such as reducing poverty, improving public health, and promoting social justice.
- Generating economic benefits : Research results can lead to the development of new products, services, and technologies that can create economic value and improve quality of life.
- Supporting academic and professional development : Research results can be used to support academic and professional development by providing opportunities for students, researchers, and practitioners to learn about new findings and methodologies in their field.
- Enhancing public understanding: Research results can help to educate the public about important issues and promote scientific literacy, leading to more informed decision-making and better public policy.
- Evaluating interventions: Research results can be used to evaluate the effectiveness of interventions, such as treatments, educational programs, and social policies. This can help to identify areas where improvements are needed and guide future interventions.
- Contributing to scientific progress: Research results can contribute to the advancement of science by providing new insights and discoveries that can lead to new theories, methods, and techniques.
- Informing decision-making : Research results can provide decision-makers with the information they need to make informed decisions. This can include decision-making at the individual, organizational, or governmental levels.
- Fostering collaboration : Research results can facilitate collaboration between researchers and practitioners, leading to new partnerships, interdisciplinary approaches, and innovative solutions to complex problems.
Advantages of Research Results
Some Advantages of Research Results are as follows:
- Improved decision-making: Research results can help inform decision-making in various fields, including medicine, business, and government. For example, research on the effectiveness of different treatments for a particular disease can help doctors make informed decisions about the best course of treatment for their patients.
- Innovation : Research results can lead to the development of new technologies, products, and services. For example, research on renewable energy sources can lead to the development of new and more efficient ways to harness renewable energy.
- Economic benefits: Research results can stimulate economic growth by providing new opportunities for businesses and entrepreneurs. For example, research on new materials or manufacturing techniques can lead to the development of new products and processes that can create new jobs and boost economic activity.
- Improved quality of life: Research results can contribute to improving the quality of life for individuals and society as a whole. For example, research on the causes of a particular disease can lead to the development of new treatments and cures, improving the health and well-being of millions of people.
About the author
Muhammad Hassan
Researcher, Academic Writer, Web developer
You may also like

How to Cite Research Paper – All Formats and...

Data Collection – Methods Types and Examples

Delimitations in Research – Types, Examples and...

Research Paper Format – Types, Examples and...

Research Process – Steps, Examples and Tips

Research Design – Types, Methods and Examples
- USC Libraries
- Research Guides
Organizing Your Social Sciences Research Paper
- 8. The Discussion
- Purpose of Guide
- Design Flaws to Avoid
- Independent and Dependent Variables
- Glossary of Research Terms
- Reading Research Effectively
- Narrowing a Topic Idea
- Broadening a Topic Idea
- Extending the Timeliness of a Topic Idea
- Academic Writing Style
- Applying Critical Thinking
- Choosing a Title
- Making an Outline
- Paragraph Development
- Research Process Video Series
- Executive Summary
- The C.A.R.S. Model
- Background Information
- The Research Problem/Question
- Theoretical Framework
- Citation Tracking
- Content Alert Services
- Evaluating Sources
- Primary Sources
- Secondary Sources
- Tiertiary Sources
- Scholarly vs. Popular Publications
- Qualitative Methods
- Quantitative Methods
- Insiderness
- Using Non-Textual Elements
- Limitations of the Study
- Common Grammar Mistakes
- Writing Concisely
- Avoiding Plagiarism
- Footnotes or Endnotes?
- Further Readings
- Generative AI and Writing
- USC Libraries Tutorials and Other Guides
- Bibliography
The purpose of the discussion section is to interpret and describe the significance of your findings in relation to what was already known about the research problem being investigated and to explain any new understanding or insights that emerged as a result of your research. The discussion will always connect to the introduction by way of the research questions or hypotheses you posed and the literature you reviewed, but the discussion does not simply repeat or rearrange the first parts of your paper; the discussion clearly explains how your study advanced the reader's understanding of the research problem from where you left them at the end of your review of prior research.
Annesley, Thomas M. “The Discussion Section: Your Closing Argument.” Clinical Chemistry 56 (November 2010): 1671-1674; Peacock, Matthew. “Communicative Moves in the Discussion Section of Research Articles.” System 30 (December 2002): 479-497.
Importance of a Good Discussion
The discussion section is often considered the most important part of your research paper because it:
- Most effectively demonstrates your ability as a researcher to think critically about an issue, to develop creative solutions to problems based upon a logical synthesis of the findings, and to formulate a deeper, more profound understanding of the research problem under investigation;
- Presents the underlying meaning of your research, notes possible implications in other areas of study, and explores possible improvements that can be made in order to further develop the concerns of your research;
- Highlights the importance of your study and how it can contribute to understanding the research problem within the field of study;
- Presents how the findings from your study revealed and helped fill gaps in the literature that had not been previously exposed or adequately described; and,
- Engages the reader in thinking critically about issues based on an evidence-based interpretation of findings; it is not governed strictly by objective reporting of information.
Annesley Thomas M. “The Discussion Section: Your Closing Argument.” Clinical Chemistry 56 (November 2010): 1671-1674; Bitchener, John and Helen Basturkmen. “Perceptions of the Difficulties of Postgraduate L2 Thesis Students Writing the Discussion Section.” Journal of English for Academic Purposes 5 (January 2006): 4-18; Kretchmer, Paul. Fourteen Steps to Writing an Effective Discussion Section. San Francisco Edit, 2003-2008.
Structure and Writing Style
I. General Rules
These are the general rules you should adopt when composing your discussion of the results :
- Do not be verbose or repetitive; be concise and make your points clearly
- Avoid the use of jargon or undefined technical language
- Follow a logical stream of thought; in general, interpret and discuss the significance of your findings in the same sequence you described them in your results section [a notable exception is to begin by highlighting an unexpected result or a finding that can grab the reader's attention]
- Use the present verb tense, especially for established facts; however, refer to specific works or prior studies in the past tense
- If needed, use subheadings to help organize your discussion or to categorize your interpretations into themes
II. The Content
The content of the discussion section of your paper most often includes :
- Explanation of results : Comment on whether or not the results were expected for each set of findings; go into greater depth to explain findings that were unexpected or especially profound. If appropriate, note any unusual or unanticipated patterns or trends that emerged from your results and explain their meaning in relation to the research problem.
- References to previous research : Either compare your results with the findings from other studies or use the studies to support a claim. This can include re-visiting key sources already cited in your literature review section, or, save them to cite later in the discussion section if they are more important to compare with your results instead of being a part of the general literature review of prior research used to provide context and background information. Note that you can make this decision to highlight specific studies after you have begun writing the discussion section.
- Deduction : A claim for how the results can be applied more generally. For example, describing lessons learned, proposing recommendations that can help improve a situation, or highlighting best practices.
- Hypothesis : A more general claim or possible conclusion arising from the results [which may be proved or disproved in subsequent research]. This can be framed as new research questions that emerged as a consequence of your analysis.
III. Organization and Structure
Keep the following sequential points in mind as you organize and write the discussion section of your paper:
- Think of your discussion as an inverted pyramid. Organize the discussion from the general to the specific, linking your findings to the literature, then to theory, then to practice [if appropriate].
- Use the same key terms, narrative style, and verb tense [present] that you used when describing the research problem in your introduction.
- Begin by briefly re-stating the research problem you were investigating and answer all of the research questions underpinning the problem that you posed in the introduction.
- Describe the patterns, principles, and relationships shown by each major findings and place them in proper perspective. The sequence of this information is important; first state the answer, then the relevant results, then cite the work of others. If appropriate, refer the reader to a figure or table to help enhance the interpretation of the data [either within the text or as an appendix].
- Regardless of where it's mentioned, a good discussion section includes analysis of any unexpected findings. This part of the discussion should begin with a description of the unanticipated finding, followed by a brief interpretation as to why you believe it appeared and, if necessary, its possible significance in relation to the overall study. If more than one unexpected finding emerged during the study, describe each of them in the order they appeared as you gathered or analyzed the data. As noted, the exception to discussing findings in the same order you described them in the results section would be to begin by highlighting the implications of a particularly unexpected or significant finding that emerged from the study, followed by a discussion of the remaining findings.
- Before concluding the discussion, identify potential limitations and weaknesses if you do not plan to do so in the conclusion of the paper. Comment on their relative importance in relation to your overall interpretation of the results and, if necessary, note how they may affect the validity of your findings. Avoid using an apologetic tone; however, be honest and self-critical [e.g., in retrospect, had you included a particular question in a survey instrument, additional data could have been revealed].
- The discussion section should end with a concise summary of the principal implications of the findings regardless of their significance. Give a brief explanation about why you believe the findings and conclusions of your study are important and how they support broader knowledge or understanding of the research problem. This can be followed by any recommendations for further research. However, do not offer recommendations which could have been easily addressed within the study. This would demonstrate to the reader that you have inadequately examined and interpreted the data.
IV. Overall Objectives
The objectives of your discussion section should include the following: I. Reiterate the Research Problem/State the Major Findings
Briefly reiterate the research problem or problems you are investigating and the methods you used to investigate them, then move quickly to describe the major findings of the study. You should write a direct, declarative, and succinct proclamation of the study results, usually in one paragraph.
II. Explain the Meaning of the Findings and Why They are Important
No one has thought as long and hard about your study as you have. Systematically explain the underlying meaning of your findings and state why you believe they are significant. After reading the discussion section, you want the reader to think critically about the results and why they are important. You don’t want to force the reader to go through the paper multiple times to figure out what it all means. If applicable, begin this part of the section by repeating what you consider to be your most significant or unanticipated finding first, then systematically review each finding. Otherwise, follow the general order you reported the findings presented in the results section.
III. Relate the Findings to Similar Studies
No study in the social sciences is so novel or possesses such a restricted focus that it has absolutely no relation to previously published research. The discussion section should relate your results to those found in other studies, particularly if questions raised from prior studies served as the motivation for your research. This is important because comparing and contrasting the findings of other studies helps to support the overall importance of your results and it highlights how and in what ways your study differs from other research about the topic. Note that any significant or unanticipated finding is often because there was no prior research to indicate the finding could occur. If there is prior research to indicate this, you need to explain why it was significant or unanticipated. IV. Consider Alternative Explanations of the Findings
It is important to remember that the purpose of research in the social sciences is to discover and not to prove . When writing the discussion section, you should carefully consider all possible explanations for the study results, rather than just those that fit your hypothesis or prior assumptions and biases. This is especially important when describing the discovery of significant or unanticipated findings.
V. Acknowledge the Study’s Limitations
It is far better for you to identify and acknowledge your study’s limitations than to have them pointed out by your professor! Note any unanswered questions or issues your study could not address and describe the generalizability of your results to other situations. If a limitation is applicable to the method chosen to gather information, then describe in detail the problems you encountered and why. VI. Make Suggestions for Further Research
You may choose to conclude the discussion section by making suggestions for further research [as opposed to offering suggestions in the conclusion of your paper]. Although your study can offer important insights about the research problem, this is where you can address other questions related to the problem that remain unanswered or highlight hidden issues that were revealed as a result of conducting your research. You should frame your suggestions by linking the need for further research to the limitations of your study [e.g., in future studies, the survey instrument should include more questions that ask..."] or linking to critical issues revealed from the data that were not considered initially in your research.
NOTE: Besides the literature review section, the preponderance of references to sources is usually found in the discussion section . A few historical references may be helpful for perspective, but most of the references should be relatively recent and included to aid in the interpretation of your results, to support the significance of a finding, and/or to place a finding within a particular context. If a study that you cited does not support your findings, don't ignore it--clearly explain why your research findings differ from theirs.
V. Problems to Avoid
- Do not waste time restating your results . Should you need to remind the reader of a finding to be discussed, use "bridge sentences" that relate the result to the interpretation. An example would be: “In the case of determining available housing to single women with children in rural areas of Texas, the findings suggest that access to good schools is important...," then move on to further explaining this finding and its implications.
- As noted, recommendations for further research can be included in either the discussion or conclusion of your paper, but do not repeat your recommendations in the both sections. Think about the overall narrative flow of your paper to determine where best to locate this information. However, if your findings raise a lot of new questions or issues, consider including suggestions for further research in the discussion section.
- Do not introduce new results in the discussion section. Be wary of mistaking the reiteration of a specific finding for an interpretation because it may confuse the reader. The description of findings [results section] and the interpretation of their significance [discussion section] should be distinct parts of your paper. If you choose to combine the results section and the discussion section into a single narrative, you must be clear in how you report the information discovered and your own interpretation of each finding. This approach is not recommended if you lack experience writing college-level research papers.
- Use of the first person pronoun is generally acceptable. Using first person singular pronouns can help emphasize a point or illustrate a contrasting finding. However, keep in mind that too much use of the first person can actually distract the reader from the main points [i.e., I know you're telling me this--just tell me!].
Analyzing vs. Summarizing. Department of English Writing Guide. George Mason University; Discussion. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. Department of Biology. Bates College; Hess, Dean R. "How to Write an Effective Discussion." Respiratory Care 49 (October 2004); Kretchmer, Paul. Fourteen Steps to Writing to Writing an Effective Discussion Section. San Francisco Edit, 2003-2008; The Lab Report. University College Writing Centre. University of Toronto; Sauaia, A. et al. "The Anatomy of an Article: The Discussion Section: "How Does the Article I Read Today Change What I Will Recommend to my Patients Tomorrow?” The Journal of Trauma and Acute Care Surgery 74 (June 2013): 1599-1602; Research Limitations & Future Research . Lund Research Ltd., 2012; Summary: Using it Wisely. The Writing Center. University of North Carolina; Schafer, Mickey S. Writing the Discussion. Writing in Psychology course syllabus. University of Florida; Yellin, Linda L. A Sociology Writer's Guide . Boston, MA: Allyn and Bacon, 2009.
Writing Tip
Don’t Over-Interpret the Results!
Interpretation is a subjective exercise. As such, you should always approach the selection and interpretation of your findings introspectively and to think critically about the possibility of judgmental biases unintentionally entering into discussions about the significance of your work. With this in mind, be careful that you do not read more into the findings than can be supported by the evidence you have gathered. Remember that the data are the data: nothing more, nothing less.
MacCoun, Robert J. "Biases in the Interpretation and Use of Research Results." Annual Review of Psychology 49 (February 1998): 259-287; Ward, Paulet al, editors. The Oxford Handbook of Expertise . Oxford, UK: Oxford University Press, 2018.
Another Writing Tip
Don't Write Two Results Sections!
One of the most common mistakes that you can make when discussing the results of your study is to present a superficial interpretation of the findings that more or less re-states the results section of your paper. Obviously, you must refer to your results when discussing them, but focus on the interpretation of those results and their significance in relation to the research problem, not the data itself.
Azar, Beth. "Discussing Your Findings." American Psychological Association gradPSYCH Magazine (January 2006).
Yet Another Writing Tip
Avoid Unwarranted Speculation!
The discussion section should remain focused on the findings of your study. For example, if the purpose of your research was to measure the impact of foreign aid on increasing access to education among disadvantaged children in Bangladesh, it would not be appropriate to speculate about how your findings might apply to populations in other countries without drawing from existing studies to support your claim or if analysis of other countries was not a part of your original research design. If you feel compelled to speculate, do so in the form of describing possible implications or explaining possible impacts. Be certain that you clearly identify your comments as speculation or as a suggestion for where further research is needed. Sometimes your professor will encourage you to expand your discussion of the results in this way, while others don’t care what your opinion is beyond your effort to interpret the data in relation to the research problem.
- << Previous: Using Non-Textual Elements
- Next: Limitations of the Study >>
- Last Updated: Apr 9, 2024 1:19 PM
- URL: https://libguides.usc.edu/writingguide
How to Write the Discussion Section of a Research Paper
The discussion section of a research paper analyzes and interprets the findings, provides context, compares them with previous studies, identifies limitations, and suggests future research directions.
Updated on September 15, 2023

Structure your discussion section right, and you’ll be cited more often while doing a greater service to the scientific community. So, what actually goes into the discussion section? And how do you write it?
The discussion section of your research paper is where you let the reader know how your study is positioned in the literature, what to take away from your paper, and how your work helps them. It can also include your conclusions and suggestions for future studies.
First, we’ll define all the parts of your discussion paper, and then look into how to write a strong, effective discussion section for your paper or manuscript.
Discussion section: what is it, what it does
The discussion section comes later in your paper, following the introduction, methods, and results. The discussion sets up your study’s conclusions. Its main goals are to present, interpret, and provide a context for your results.
What is it?
The discussion section provides an analysis and interpretation of the findings, compares them with previous studies, identifies limitations, and suggests future directions for research.
This section combines information from the preceding parts of your paper into a coherent story. By this point, the reader already knows why you did your study (introduction), how you did it (methods), and what happened (results). In the discussion, you’ll help the reader connect the ideas from these sections.
Why is it necessary?
The discussion provides context and interpretations for the results. It also answers the questions posed in the introduction. While the results section describes your findings, the discussion explains what they say. This is also where you can describe the impact or implications of your research.
Adds context for your results
Most research studies aim to answer a question, replicate a finding, or address limitations in the literature. These goals are first described in the introduction. However, in the discussion section, the author can refer back to them to explain how the study's objective was achieved.
Shows what your results actually mean and real-world implications
The discussion can also describe the effect of your findings on research or practice. How are your results significant for readers, other researchers, or policymakers?
What to include in your discussion (in the correct order)
A complete and effective discussion section should at least touch on the points described below.
Summary of key findings
The discussion should begin with a brief factual summary of the results. Concisely overview the main results you obtained.
Begin with key findings with supporting evidence
Your results section described a list of findings, but what message do they send when you look at them all together?
Your findings were detailed in the results section, so there’s no need to repeat them here, but do provide at least a few highlights. This will help refresh the reader’s memory and help them focus on the big picture.
Read the first paragraph of the discussion section in this article (PDF) for an example of how to start this part of your paper. Notice how the authors break down their results and follow each description sentence with an explanation of why each finding is relevant.
State clearly and concisely
Following a clear and direct writing style is especially important in the discussion section. After all, this is where you will make some of the most impactful points in your paper. While the results section often contains technical vocabulary, such as statistical terms, the discussion section lets you describe your findings more clearly.
Interpretation of results
Once you’ve given your reader an overview of your results, you need to interpret those results. In other words, what do your results mean? Discuss the findings’ implications and significance in relation to your research question or hypothesis.
Analyze and interpret your findings
Look into your findings and explore what’s behind them or what may have caused them. If your introduction cited theories or studies that could explain your findings, use these sources as a basis to discuss your results.
For example, look at the second paragraph in the discussion section of this article on waggling honey bees. Here, the authors explore their results based on information from the literature.
Unexpected or contradictory results
Sometimes, your findings are not what you expect. Here’s where you describe this and try to find a reason for it. Could it be because of the method you used? Does it have something to do with the variables analyzed? Comparing your methods with those of other similar studies can help with this task.
Context and comparison with previous work
Refer to related studies to place your research in a larger context and the literature. Compare and contrast your findings with existing literature, highlighting similarities, differences, and/or contradictions.
How your work compares or contrasts with previous work
Studies with similar findings to yours can be cited to show the strength of your findings. Information from these studies can also be used to help explain your results. Differences between your findings and others in the literature can also be discussed here.
How to divide this section into subsections
If you have more than one objective in your study or many key findings, you can dedicate a separate section to each of these. Here’s an example of this approach. You can see that the discussion section is divided into topics and even has a separate heading for each of them.
Limitations
Many journals require you to include the limitations of your study in the discussion. Even if they don’t, there are good reasons to mention these in your paper.
Why limitations don’t have a negative connotation
A study’s limitations are points to be improved upon in future research. While some of these may be flaws in your method, many may be due to factors you couldn’t predict.
Examples include time constraints or small sample sizes. Pointing this out will help future researchers avoid or address these issues. This part of the discussion can also include any attempts you have made to reduce the impact of these limitations, as in this study .
How limitations add to a researcher's credibility
Pointing out the limitations of your study demonstrates transparency. It also shows that you know your methods well and can conduct a critical assessment of them.
Implications and significance
The final paragraph of the discussion section should contain the take-home messages for your study. It can also cite the “strong points” of your study, to contrast with the limitations section.
Restate your hypothesis
Remind the reader what your hypothesis was before you conducted the study.
How was it proven or disproven?
Identify your main findings and describe how they relate to your hypothesis.
How your results contribute to the literature
Were you able to answer your research question? Or address a gap in the literature?
Future implications of your research
Describe the impact that your results may have on the topic of study. Your results may show, for instance, that there are still limitations in the literature for future studies to address. There may be a need for studies that extend your findings in a specific way. You also may need additional research to corroborate your findings.
Sample discussion section
This fictitious example covers all the aspects discussed above. Your actual discussion section will probably be much longer, but you can read this to get an idea of everything your discussion should cover.
Our results showed that the presence of cats in a household is associated with higher levels of perceived happiness by its human occupants. These findings support our hypothesis and demonstrate the association between pet ownership and well-being.
The present findings align with those of Bao and Schreer (2016) and Hardie et al. (2023), who observed greater life satisfaction in pet owners relative to non-owners. Although the present study did not directly evaluate life satisfaction, this factor may explain the association between happiness and cat ownership observed in our sample.
Our findings must be interpreted in light of some limitations, such as the focus on cat ownership only rather than pets as a whole. This may limit the generalizability of our results.
Nevertheless, this study had several strengths. These include its strict exclusion criteria and use of a standardized assessment instrument to investigate the relationships between pets and owners. These attributes bolster the accuracy of our results and reduce the influence of confounding factors, increasing the strength of our conclusions. Future studies may examine the factors that mediate the association between pet ownership and happiness to better comprehend this phenomenon.
This brief discussion begins with a quick summary of the results and hypothesis. The next paragraph cites previous research and compares its findings to those of this study. Information from previous studies is also used to help interpret the findings. After discussing the results of the study, some limitations are pointed out. The paper also explains why these limitations may influence the interpretation of results. Then, final conclusions are drawn based on the study, and directions for future research are suggested.
How to make your discussion flow naturally
If you find writing in scientific English challenging, the discussion and conclusions are often the hardest parts of the paper to write. That’s because you’re not just listing up studies, methods, and outcomes. You’re actually expressing your thoughts and interpretations in words.
- How formal should it be?
- What words should you use, or not use?
- How do you meet strict word limits, or make it longer and more informative?
Always give it your best, but sometimes a helping hand can, well, help. Getting a professional edit can help clarify your work’s importance while improving the English used to explain it. When readers know the value of your work, they’ll cite it. We’ll assign your study to an expert editor knowledgeable in your area of research. Their work will clarify your discussion, helping it to tell your story. Find out more about AJE Editing.

Adam Goulston, PsyD, MS, MBA, MISD, ELS
Science Marketing Consultant
See our "Privacy Policy"
Ensure your structure and ideas are consistent and clearly communicated
Pair your Premium Editing with our add-on service Presubmission Review for an overall assessment of your manuscript.

- Langson Library
- Science Library
- Grunigen Medical Library
- Law Library
- Connect From Off-Campus
- Accessibility
- Gateway Study Center

Email this link
Writing a scientific paper.
- Writing a lab report
- INTRODUCTION
Writing a "good" discussion section
"discussion and conclusions checklist" from: how to write a good scientific paper. chris a. mack. spie. 2018., peer review.
- LITERATURE CITED
- Bibliography of guides to scientific writing and presenting
- Presentations
- Lab Report Writing Guides on the Web
This is is usually the hardest section to write. You are trying to bring out the true meaning of your data without being too long. Do not use words to conceal your facts or reasoning. Also do not repeat your results, this is a discussion.
- Present principles, relationships and generalizations shown by the results
- Point out exceptions or lack of correlations. Define why you think this is so.
- Show how your results agree or disagree with previously published works
- Discuss the theoretical implications of your work as well as practical applications
- State your conclusions clearly. Summarize your evidence for each conclusion.
- Discuss the significance of the results
- Evidence does not explain itself; the results must be presented and then explained.
- Typical stages in the discussion: summarizing the results, discussing whether results are expected or unexpected, comparing these results to previous work, interpreting and explaining the results (often by comparison to a theory or model), and hypothesizing about their generality.
- Discuss any problems or shortcomings encountered during the course of the work.
- Discuss possible alternate explanations for the results.
- Avoid: presenting results that are never discussed; presenting discussion that does not relate to any of the results; presenting results and discussion in chronological order rather than logical order; ignoring results that do not support the conclusions; drawing conclusions from results without logical arguments to back them up.
CONCLUSIONS
- Provide a very brief summary of the Results and Discussion.
- Emphasize the implications of the findings, explaining how the work is significant and providing the key message(s) the author wishes to convey.
- Provide the most general claims that can be supported by the evidence.
- Provide a future perspective on the work.
- Avoid: repeating the abstract; repeating background information from the Introduction; introducing new evidence or new arguments not found in the Results and Discussion; repeating the arguments made in the Results and Discussion; failing to address all of the research questions set out in the Introduction.
WHAT HAPPENS AFTER I COMPLETE MY PAPER?
The peer review process is the quality control step in the publication of ideas. Papers that are submitted to a journal for publication are sent out to several scientists (peers) who look carefully at the paper to see if it is "good science". These reviewers then recommend to the editor of a journal whether or not a paper should be published. Most journals have publication guidelines. Ask for them and follow them exactly. Peer reviewers examine the soundness of the materials and methods section. Are the materials and methods used written clearly enough for another scientist to reproduce the experiment? Other areas they look at are: originality of research, significance of research question studied, soundness of the discussion and interpretation, correct spelling and use of technical terms, and length of the article.
- << Previous: RESULTS
- Next: LITERATURE CITED >>
- Last Updated: Aug 4, 2023 9:33 AM
- URL: https://guides.lib.uci.edu/scientificwriting
Off-campus? Please use the Software VPN and choose the group UCIFull to access licensed content. For more information, please Click here
Software VPN is not available for guests, so they may not have access to some content when connecting from off-campus.
- Affiliate Program

- UNITED STATES
- 台灣 (TAIWAN)
- TÜRKIYE (TURKEY)
- Academic Editing Services
- - Research Paper
- - Journal Manuscript
- - Dissertation
- - College & University Assignments
- Admissions Editing Services
- - Application Essay
- - Personal Statement
- - Recommendation Letter
- - Cover Letter
- - CV/Resume
- Business Editing Services
- - Business Documents
- - Report & Brochure
- - Website & Blog
- Writer Editing Services
- - Script & Screenplay
- Our Editors
- Client Reviews
- Editing & Proofreading Prices
- Wordvice Points
- Partner Discount
- Plagiarism Checker
- APA Citation Generator
- MLA Citation Generator
- Chicago Citation Generator
- Vancouver Citation Generator
- - APA Style
- - MLA Style
- - Chicago Style
- - Vancouver Style
- Writing & Editing Guide
- Academic Resources
- Admissions Resources
How to Write a Discussion Section for a Research Paper
We’ve talked about several useful writing tips that authors should consider while drafting or editing their research papers. In particular, we’ve focused on figures and legends , as well as the Introduction , Methods , and Results . Now that we’ve addressed the more technical portions of your journal manuscript, let’s turn to the analytical segments of your research article. In this article, we’ll provide tips on how to write a strong Discussion section that best portrays the significance of your research contributions.
What is the Discussion section of a research paper?
In a nutshell, your Discussion fulfills the promise you made to readers in your Introduction . At the beginning of your paper, you tell us why we should care about your research. You then guide us through a series of intricate images and graphs that capture all the relevant data you collected during your research. We may be dazzled and impressed at first, but none of that matters if you deliver an anti-climactic conclusion in the Discussion section!
Are you feeling pressured? Don’t worry. To be honest, you will edit the Discussion section of your manuscript numerous times. After all, in as little as one to two paragraphs ( Nature ‘s suggestion based on their 3,000-word main body text limit), you have to explain how your research moves us from point A (issues you raise in the Introduction) to point B (our new understanding of these matters). You must also recommend how we might get to point C (i.e., identify what you think is the next direction for research in this field). That’s a lot to say in two paragraphs!
So, how do you do that? Let’s take a closer look.
What should I include in the Discussion section?
As we stated above, the goal of your Discussion section is to answer the questions you raise in your Introduction by using the results you collected during your research . The content you include in the Discussions segment should include the following information:
- Remind us why we should be interested in this research project.
- Describe the nature of the knowledge gap you were trying to fill using the results of your study.
- Don’t repeat your Introduction. Instead, focus on why this particular study was needed to fill the gap you noticed and why that gap needed filling in the first place.
- Mainly, you want to remind us of how your research will increase our knowledge base and inspire others to conduct further research.
- Clearly tell us what that piece of missing knowledge was.
- Answer each of the questions you asked in your Introduction and explain how your results support those conclusions.
- Make sure to factor in all results relevant to the questions (even if those results were not statistically significant).
- Focus on the significance of the most noteworthy results.
- If conflicting inferences can be drawn from your results, evaluate the merits of all of them.
- Don’t rehash what you said earlier in the Results section. Rather, discuss your findings in the context of answering your hypothesis. Instead of making statements like “[The first result] was this…,” say, “[The first result] suggests [conclusion].”
- Do your conclusions line up with existing literature?
- Discuss whether your findings agree with current knowledge and expectations.
- Keep in mind good persuasive argument skills, such as explaining the strengths of your arguments and highlighting the weaknesses of contrary opinions.
- If you discovered something unexpected, offer reasons. If your conclusions aren’t aligned with current literature, explain.
- Address any limitations of your study and how relevant they are to interpreting your results and validating your findings.
- Make sure to acknowledge any weaknesses in your conclusions and suggest room for further research concerning that aspect of your analysis.
- Make sure your suggestions aren’t ones that should have been conducted during your research! Doing so might raise questions about your initial research design and protocols.
- Similarly, maintain a critical but unapologetic tone. You want to instill confidence in your readers that you have thoroughly examined your results and have objectively assessed them in a way that would benefit the scientific community’s desire to expand our knowledge base.
- Recommend next steps.
- Your suggestions should inspire other researchers to conduct follow-up studies to build upon the knowledge you have shared with them.
- Keep the list short (no more than two).
How to Write the Discussion Section
The above list of what to include in the Discussion section gives an overall idea of what you need to focus on throughout the section. Below are some tips and general suggestions about the technical aspects of writing and organization that you might find useful as you draft or revise the contents we’ve outlined above.
Technical writing elements
- Embrace active voice because it eliminates the awkward phrasing and wordiness that accompanies passive voice.
- Use the present tense, which should also be employed in the Introduction.
- Sprinkle with first person pronouns if needed, but generally, avoid it. We want to focus on your findings.
- Maintain an objective and analytical tone.
Discussion section organization
- Keep the same flow across the Results, Methods, and Discussion sections.
- We develop a rhythm as we read and parallel structures facilitate our comprehension. When you organize information the same way in each of these related parts of your journal manuscript, we can quickly see how a certain result was interpreted and quickly verify the particular methods used to produce that result.
- Notice how using parallel structure will eliminate extra narration in the Discussion part since we can anticipate the flow of your ideas based on what we read in the Results segment. Reducing wordiness is important when you only have a few paragraphs to devote to the Discussion section!
- Within each subpart of a Discussion, the information should flow as follows: (A) conclusion first, (B) relevant results and how they relate to that conclusion and (C) relevant literature.
- End with a concise summary explaining the big-picture impact of your study on our understanding of the subject matter. At the beginning of your Discussion section, you stated why this particular study was needed to fill the gap you noticed and why that gap needed filling in the first place. Now, it is time to end with “how your research filled that gap.”
Discussion Part 1: Summarizing Key Findings
Begin the Discussion section by restating your statement of the problem and briefly summarizing the major results. Do not simply repeat your findings. Rather, try to create a concise statement of the main results that directly answer the central research question that you stated in the Introduction section . This content should not be longer than one paragraph in length.
Many researchers struggle with understanding the precise differences between a Discussion section and a Results section . The most important thing to remember here is that your Discussion section should subjectively evaluate the findings presented in the Results section, and in relatively the same order. Keep these sections distinct by making sure that you do not repeat the findings without providing an interpretation.
Phrase examples: Summarizing the results
- The findings indicate that …
- These results suggest a correlation between A and B …
- The data present here suggest that …
- An interpretation of the findings reveals a connection between…
Discussion Part 2: Interpreting the Findings
What do the results mean? It may seem obvious to you, but simply looking at the figures in the Results section will not necessarily convey to readers the importance of the findings in answering your research questions.
The exact structure of interpretations depends on the type of research being conducted. Here are some common approaches to interpreting data:
- Identifying correlations and relationships in the findings
- Explaining whether the results confirm or undermine your research hypothesis
- Giving the findings context within the history of similar research studies
- Discussing unexpected results and analyzing their significance to your study or general research
- Offering alternative explanations and arguing for your position
Organize the Discussion section around key arguments, themes, hypotheses, or research questions or problems. Again, make sure to follow the same order as you did in the Results section.
Discussion Part 3: Discussing the Implications
In addition to providing your own interpretations, show how your results fit into the wider scholarly literature you surveyed in the literature review section. This section is called the implications of the study . Show where and how these results fit into existing knowledge, what additional insights they contribute, and any possible consequences that might arise from this knowledge, both in the specific research topic and in the wider scientific domain.
Questions to ask yourself when dealing with potential implications:
- Do your findings fall in line with existing theories, or do they challenge these theories or findings? What new information do they contribute to the literature, if any? How exactly do these findings impact or conflict with existing theories or models?
- What are the practical implications on actual subjects or demographics?
- What are the methodological implications for similar studies conducted either in the past or future?
Your purpose in giving the implications is to spell out exactly what your study has contributed and why researchers and other readers should be interested.
Phrase examples: Discussing the implications of the research
- These results confirm the existing evidence in X studies…
- The results are not in line with the foregoing theory that…
- This experiment provides new insights into the connection between…
- These findings present a more nuanced understanding of…
- While previous studies have focused on X, these results demonstrate that Y.
Step 4: Acknowledging the limitations
All research has study limitations of one sort or another. Acknowledging limitations in methodology or approach helps strengthen your credibility as a researcher. Study limitations are not simply a list of mistakes made in the study. Rather, limitations help provide a more detailed picture of what can or cannot be concluded from your findings. In essence, they help temper and qualify the study implications you listed previously.
Study limitations can relate to research design, specific methodological or material choices, or unexpected issues that emerged while you conducted the research. Mention only those limitations directly relate to your research questions, and explain what impact these limitations had on how your study was conducted and the validity of any interpretations.
Possible types of study limitations:
- Insufficient sample size for statistical measurements
- Lack of previous research studies on the topic
- Methods/instruments/techniques used to collect the data
- Limited access to data
- Time constraints in properly preparing and executing the study
After discussing the study limitations, you can also stress that your results are still valid. Give some specific reasons why the limitations do not necessarily handicap your study or narrow its scope.
Phrase examples: Limitations sentence beginners
- “There may be some possible limitations in this study.”
- “The findings of this study have to be seen in light of some limitations.”
- “The first limitation is the…The second limitation concerns the…”
- “The empirical results reported herein should be considered in the light of some limitations.”
- “This research, however, is subject to several limitations.”
- “The primary limitation to the generalization of these results is…”
- “Nonetheless, these results must be interpreted with caution and a number of limitations should be borne in mind.”
Discussion Part 5: Giving Recommendations for Further Research
Based on your interpretation and discussion of the findings, your recommendations can include practical changes to the study or specific further research to be conducted to clarify the research questions. Recommendations are often listed in a separate Conclusion section , but often this is just the final paragraph of the Discussion section.
Suggestions for further research often stem directly from the limitations outlined. Rather than simply stating that “further research should be conducted,” provide concrete specifics for how future can help answer questions that your research could not.
Phrase examples: Recommendation sentence beginners
- Further research is needed to establish …
- There is abundant space for further progress in analyzing…
- A further study with more focus on X should be done to investigate…
- Further studies of X that account for these variables must be undertaken.
Consider Receiving Professional Language Editing
As you edit or draft your research manuscript, we hope that you implement these guidelines to produce a more effective Discussion section. And after completing your draft, don’t forget to submit your work to a professional proofreading and English editing service like Wordvice, including our manuscript editing service for paper editing , cover letter editing , SOP editing , and personal statement proofreading services. Language editors not only proofread and correct errors in grammar, punctuation, mechanics, and formatting but also improve terms and revise phrases so they read more naturally. Wordvice is an industry leader in providing high-quality revision for all types of academic documents.
For additional information about how to write a strong research paper, make sure to check out our full research writing series !
Wordvice Writing Resources
- How to Write a Research Paper Introduction
- Which Verb Tenses to Use in a Research Paper
- How to Write an Abstract for a Research Paper
- How to Write a Research Paper Title
- Useful Phrases for Academic Writing
- Common Transition Terms in Academic Papers
- Active and Passive Voice in Research Papers
- 100+ Verbs That Will Make Your Research Writing Amazing
- Tips for Paraphrasing in Research Papers
Additional Academic Resources
- Guide for Authors. (Elsevier)
- How to Write the Results Section of a Research Paper. (Bates College)
- Structure of a Research Paper. (University of Minnesota Biomedical Library)
- How to Choose a Target Journal (Springer)
- How to Write Figures and Tables (UNC Writing Center)
Training videos | Faqs


Discussion Section Examples and Writing Tips
Abstract | Introduction | Literature Review | Research question | Materials & Methods | Results | Discussion | Conclusion
In this blog, we look at how to write the discussion section of a research paper. We will go through plenty of discussion examples and understand how to construct a great discussion section for your research paper.
1. What is the purpose of the discussion section?
The discussion section is one of the most important sections of your research paper. This is where you interpret your results, highlight your contributions, and explain the value of your work to your readers. This is one of the challenging parts to write because the author must clearly explain the significance of their results and tie everything back to the research questions.
2. How should I structure my discussion section?
Generally, the discussion section of a research paper typically contains the following parts.
Research summary It is a good idea to start this section with an overall summary of your work and highlight the main findings of your research.
Interpretation of findings You must interpret your findings clearly to your readers one by one.
Comparison with literature You must talk about how your results fit into existing research in the literature.
Implications of your work You should talk about the implications and possible benefits of your research.
Limitations You should talk about the possible limitations and shortcomings of your research
Future work And finally, you can talk about the possible future directions of your work.
3. Discussion Examples
Let’s look at some examples of the discussion section. We will be looking at discussion examples from different fields and of different formats. We have split this section into multiple components so that it is easy for you to digest and understand.
3.1. An example of research summary in discussion
It is a good idea to start your discussion section with the summary of your work. The best way to do this will be to restate your research question, and then reminding your readers about your methods, and finally providing an overall summary of your results.
Our aims were to compare the effectiveness and user-friendliness of different storm detection software for storm tracking. On the basis of these aims, we ran multiple experiments with the same conditions using different storm detection software. Our results showed that in both speed and accuracy of data, ‘software A’ performed better than ‘software B’. _ Aims summary _ Methodology summary _ Results summary
This discussion example is from an engineering research paper. The authors are restating their aims first, which is to compare different types of storm-tracking software. Then, they are providing a brief summary of the methods. Here, they are testing different storm-tracking software under different conditions to see which performs the best. Then, they are finally providing their main finding which is that they found ‘software A’ better than ‘software B’. This is a very good example of how to start the discussion section by presenting a summary of your work.
3.2. An example of result interpretation in discussion
The next step is to interpret your results. You have to explain your results clearly to your readers. Here is a discussion example that shows how to interpret your results.
The results of this study indicate significant differences between classical music and pop music in terms of their effects on memory recall and cognition. This implies that as the complexity of the music increases, so does its ability to facilitate cognitive processing. This finding aligns with the well-known “Mozart effect,” which suggests that listening to classical music can enhance cognitive function. _ Result _ Interpretation _ Additional evidence
The authors are saying that their results show that there is a significant difference between pop music and classical music in terms of memory recall and cognition. Now they are providing their interpretation of the findings. They think it is because there is a link between the complexity of music and cognitive processing. They are also making a reference to a well-known theory called the ‘Mozart effect’ to back up their findings. It is a nicely written passage and the author’s interpretation sounds very convincing and credible.
3.3. An example of literature comparison in discussion
The next step is to compare your results to the literature. You have to explain clearly how your findings compare with similar findings made by other researchers. Here is a discussion example where authors are providing details of papers in the literature that both support and oppose their findings.
Our analysis predicts that climate change will have a significant impact on wheat yield. This finding undermines one of the central pieces of evidence in some previous simulation studies [1-3] that suggest a negative effect of climate change on wheat yield, but the result is entirely consistent with the predictions of other research [4-5] that suggests the overall change in climate could result in increases in wheat yield. _ Result _ Comparison with literature
The authors are saying that their results show that climate change will have a significant effect on wheat production. Then, they are saying that there are some papers in the literature that are in agreement with their findings. However, there are also many papers in the literature that disagree with their findings. This is very important. Your discussion should be two-sided, not one-sided. You should not ignore the literature that doesn’t corroborate your findings.
3.4. An example of research implications in discussion
The next step is to explain to your readers how your findings will benefit society and the research community. You have to clearly explain the value of your work to your readers. Here is a discussion example where authors explain the implications of their research.
The results contribute insights with regard to the management of wildfire events using artificial intelligence. One could easily argue that the obvious practical implication of this study is that it proposes utilizing cloud-based machine vision to detect wildfires in real-time, even before the first responders receive emergency calls. _ Your finding _ Implications of your finding
In this paper, the authors are saying that their findings indicate that Artificial intelligence can be used to effectively manage wildfire events. Then, they are talking about the practical implications of their study. They are saying that their work has proven that machine learning can be used to detect wildfires in real-time. This is a great practical application and can save thousands of lives. As you can see, after reading this passage, you can immediately understand the value and significance of the work.
3.5. An example of limitations in discussion
It is very important that you discuss the limitations of your study. Limitations are flaws and shortcomings of your study. You have to tell your readers how your limitations might influence the outcomes and conclusions of your research. Most studies will have some form of limitation. So be honest and don’t hide your limitations. In reality, your readers and reviewers will be impressed with your paper if you are upfront about your limitations.
Study design and small sample size are important limitations. This could have led to an overestimation of the effect. Future research should reconfirm these findings by conducting larger-scale studies. _ Limitation _ How it might affect the results? _ How to fix the limitation?
Here is a discussion example where the author talks about study limitations. The authors are saying that the main limitations of the study are the small sample size and weak study design. Then they explain how this might have affected their results. They are saying that it is possible that they are overestimating the actual effect they are measuring. Then finally they are telling the readers that more studies with larger sample sizes should be conducted to reconfirm the findings.
As you can see, the authors are clearly explaining three things here:
3.6. An example of future work in discussion
It is important to remember not to end your paper with limitations. Finish your paper on a positive note by telling your readers about the benefits of your research and possible future directions. Here is a discussion example where the author talks about future work.
Our study highlights useful insights about the potential of biomass as a renewable energy source. Future research can extend this research in several ways, including research on how to tackle challenges that hinder the sustainability of renewable energy sources towards climate change mitigation, such as market failures, lack of information and access to raw materials. _ Benefits of your work _ Future work
The authors are starting the final paragraph of the discussion section by highlighting the benefit of their work which is the use of biomass as a renewable source of energy. Then they talk about future research. They are saying that future research can focus on how to improve the sustainability of biomass production. This is a very good example of how to finish the discussion section of your paper on a positive note.
4. Frequently Asked Questions
Sometimes you will have negative or unexpected results in your paper. You have to talk about it in your discussion section. A lot of students find it difficult to write this part. The best way to handle this situation is not to look at results as either positive or negative. A result is a result, and you will always have something important and interesting to say about your findings. Just spend some time investigating what might have caused this result and tell your readers about it.
You must talk about the limitations of your work in the discussion section of the paper. One of the important qualities that the scientific community expects from a researcher is honesty and admitting when they have made a mistake. The important trick you have to learn while presenting your limitations is to present them in a constructive way rather than being too negative about them. You must try to use positive language even when you are talking about major limitations of your work.
If you have something exciting to say about your results or found something new that nobody else has found before, then, don’t be modest and use flat language when presenting this in the discussion. Use words like ‘break through’, ‘indisputable evidence’, ‘exciting proposition’ to increase the impact of your findings.
Important thing to remember is not to overstate your findings. If you found something really interesting but are not 100% sure, you must not mislead your readers. The best way to do this will be to use words like ‘it appears’ and ‘it seems’. This will tell the readers that there is a slight possibility that you might be wrong.
Similar Posts

Writing a Questionnaire Survey Research Paper – Example & Format
In this blog, we will explain how to write a survey questionnaire paper and discuss all the important points to consider while writing the research paper.

Useful Phrases and Sentences for Academic & Research Paper Writing
In this blog, we explain various sections of a research paper and give you an overview of what these sections should contain.

Research Paper Structure – Main Sections and Parts of a Research Paper
In this blog, we explain various topics and sub-topics to be included under each section of a research paper via word cloud diagrams.

How to Write a Research Paper? A Beginners Guide with Useful Academic Phrases
This blog explains how to write a research paper and provides writing ideas in the form of academic phrases.

Writing a Medical Clinical Trial Research Paper – Example & Format
In this blog, we will teach you step-by-step how to write a clinical trial research paper for publication in a high quality scientific journal.

Abstract Section Examples and Writing Tips
In this blog, we will go through many abstract examples and understand how to construct a good abstract for your research paper.
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- 2 Share Facebook
- 5 Share Twitter
- 4 Share LinkedIn
- 6 Share Email
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Dissertation
- How to Write a Discussion Section | Tips & Examples
How to Write a Discussion Section | Tips & Examples
Published on 21 August 2022 by Shona McCombes . Revised on 25 October 2022.

The discussion section is where you delve into the meaning, importance, and relevance of your results .
It should focus on explaining and evaluating what you found, showing how it relates to your literature review , and making an argument in support of your overall conclusion . It should not be a second results section .
There are different ways to write this section, but you can focus your writing around these key elements:
- Summary: A brief recap of your key results
- Interpretations: What do your results mean?
- Implications: Why do your results matter?
- Limitations: What can’t your results tell us?
- Recommendations: Avenues for further studies or analyses
Instantly correct all language mistakes in your text
Be assured that you'll submit flawless writing. Upload your document to correct all your mistakes.

Table of contents
What not to include in your discussion section, step 1: summarise your key findings, step 2: give your interpretations, step 3: discuss the implications, step 4: acknowledge the limitations, step 5: share your recommendations, discussion section example.
There are a few common mistakes to avoid when writing the discussion section of your paper.
- Don’t introduce new results: You should only discuss the data that you have already reported in your results section .
- Don’t make inflated claims: Avoid overinterpretation and speculation that isn’t directly supported by your data.
- Don’t undermine your research: The discussion of limitations should aim to strengthen your credibility, not emphasise weaknesses or failures.
Prevent plagiarism, run a free check.
Start this section by reiterating your research problem and concisely summarising your major findings. Don’t just repeat all the data you have already reported – aim for a clear statement of the overall result that directly answers your main research question . This should be no more than one paragraph.
Many students struggle with the differences between a discussion section and a results section . The crux of the matter is that your results sections should present your results, and your discussion section should subjectively evaluate them. Try not to blend elements of these two sections, in order to keep your paper sharp.
- The results indicate that …
- The study demonstrates a correlation between …
- This analysis supports the theory that …
- The data suggest that …
The meaning of your results may seem obvious to you, but it’s important to spell out their significance for your reader, showing exactly how they answer your research question.
The form of your interpretations will depend on the type of research, but some typical approaches to interpreting the data include:
- Identifying correlations , patterns, and relationships among the data
- Discussing whether the results met your expectations or supported your hypotheses
- Contextualising your findings within previous research and theory
- Explaining unexpected results and evaluating their significance
- Considering possible alternative explanations and making an argument for your position
You can organise your discussion around key themes, hypotheses, or research questions, following the same structure as your results section. Alternatively, you can also begin by highlighting the most significant or unexpected results.
- In line with the hypothesis …
- Contrary to the hypothesised association …
- The results contradict the claims of Smith (2007) that …
- The results might suggest that x . However, based on the findings of similar studies, a more plausible explanation is x .
As well as giving your own interpretations, make sure to relate your results back to the scholarly work that you surveyed in the literature review . The discussion should show how your findings fit with existing knowledge, what new insights they contribute, and what consequences they have for theory or practice.
Ask yourself these questions:
- Do your results support or challenge existing theories? If they support existing theories, what new information do they contribute? If they challenge existing theories, why do you think that is?
- Are there any practical implications?
Your overall aim is to show the reader exactly what your research has contributed, and why they should care.
- These results build on existing evidence of …
- The results do not fit with the theory that …
- The experiment provides a new insight into the relationship between …
- These results should be taken into account when considering how to …
- The data contribute a clearer understanding of …
- While previous research has focused on x , these results demonstrate that y .
Even the best research has its limitations. Acknowledging these is important to demonstrate your credibility. Limitations aren’t about listing your errors, but about providing an accurate picture of what can and cannot be concluded from your study.
Limitations might be due to your overall research design, specific methodological choices , or unanticipated obstacles that emerged during your research process.
Here are a few common possibilities:
- If your sample size was small or limited to a specific group of people, explain how generalisability is limited.
- If you encountered problems when gathering or analysing data, explain how these influenced the results.
- If there are potential confounding variables that you were unable to control, acknowledge the effect these may have had.
After noting the limitations, you can reiterate why the results are nonetheless valid for the purpose of answering your research question.
- The generalisability of the results is limited by …
- The reliability of these data is impacted by …
- Due to the lack of data on x , the results cannot confirm …
- The methodological choices were constrained by …
- It is beyond the scope of this study to …
Based on the discussion of your results, you can make recommendations for practical implementation or further research. Sometimes, the recommendations are saved for the conclusion .
Suggestions for further research can lead directly from the limitations. Don’t just state that more studies should be done – give concrete ideas for how future work can build on areas that your own research was unable to address.
- Further research is needed to establish …
- Future studies should take into account …
- Avenues for future research include …

Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the ‘Cite this Scribbr article’ button to automatically add the citation to our free Reference Generator.
McCombes, S. (2022, October 25). How to Write a Discussion Section | Tips & Examples. Scribbr. Retrieved 9 April 2024, from https://www.scribbr.co.uk/thesis-dissertation/discussion/
Is this article helpful?
Shona McCombes
Other students also liked, how to write a results section | tips & examples, research paper appendix | example & templates, how to write a thesis or dissertation introduction.
- Research Paper Guides
- Basics of Research Paper Writing
How to Write a Discussion Section: Writing Guide
- Speech Topics
- Basics of Essay Writing
- Essay Topics
- Other Essays
- Main Academic Essays
- Research Paper Topics
- Miscellaneous
- Chicago/ Turabian
- Data & Statistics
- Methodology
- Admission Writing Tips
- Admission Advice
- Other Guides
- Student Life
- Studying Tips
- Understanding Plagiarism
- Academic Writing Tips
- Basics of Dissertation & Thesis Writing
- Essay Guides
- Formatting Guides
- Basics of Research Process
- Admission Guides
- Dissertation & Thesis Guides

Table of contents
Use our free Readability checker
The discussion section of a research paper is where the author analyzes and explains the importance of the study's results. It presents the conclusions drawn from the study, compares them to previous research, and addresses any potential limitations or weaknesses. The discussion section should also suggest areas for future research.
Everything is not that complicated if you know where to find the required information. We’ll tell you everything there is to know about writing your discussion. Our easy guide covers all important bits, including research questions and your research results. Do you know how all enumerated events are connected? Well, you will after reading this guide we’ve prepared for you!
What Is in the Discussion Section of a Research Paper
The discussion section of a research paper can be viewed as something similar to the conclusion of your paper. But not literal, of course. It’s an ultimate section where you can talk about the findings of your study. Think about these questions when writing:
- Did you answer all of the promised research questions?
- Did you mention why your work matters?
- What are your findings, and why should anyone even care?
- Does your study have a literature review?
So, answer your questions, provide proof, and don’t forget about your promises from the introduction.
How to Write a Discussion Section in 5 Steps
How to write the discussion section of a research paper is something everyone googles eventually. It's just life. But why not make everything easier? In brief, this section we’re talking about must include all following parts:
- Answers for research questions
- Literature review
- Results of the work
- Limitations of one’s study
- Overall conclusion
Indeed, all those parts may confuse anyone. So by looking at our guide, you'll save yourself some hassle. P.S. All our steps are easy and explained in detail! But if you are looking for the most efficient solution, consider using professional help. Leave your “ write my research paper for me ” order at StudyCrumb and get a customized study tailored to your requirements.
Step 1. Start Strong: Discussion Section of a Research Paper
First and foremost, how to start the discussion section of a research paper? Here’s what you should definitely consider before settling down to start writing:
- All essays or papers must begin strong. All readers will not wait for any writer to get to the point. We advise summarizing the paper's main findings.
- Moreover, you should relate both discussion and literature review to what you have discovered. Mentioning that would be a plus too.
- Make sure that an introduction or start per se is clear and concise. Word count might be needed for school. But any paper should be understandable and not too diluted.
Step 2. Answer the Questions in Your Discussion Section of a Research Paper
Writing the discussion section of a research paper also involves mentioning your questions. Remember that in your introduction, you have promised your readers to answer certain questions. Well, now it’s a perfect time to finally give the awaited answer. You need to explain all possible correlations between your findings, research questions, and literature proposed. You already had hypotheses. So were they correct, or maybe you want to propose certain corrections? Section’s main goal is to avoid open ends. It’s not a story or a fairytale with an intriguing ending. If you have several questions, you must answer them. As simple as that.
Step 3. Relate Your Results in a Discussion Section
Writing a discussion section of a research paper also requires any writer to explain their results. You will undoubtedly include an impactful literature review. However, your readers should not just try and struggle with understanding what are some specific relationships behind previous studies and your results. Your results should sound something like: “This guy in their paper discovered that apples are green. Nevertheless, I have proven via experimentation and research that apples are actually red.” Please, don’t take these results directly. It’s just an initial hypothesis. But what you should definitely remember is any practical implications of your study. Why does it matter and how can anyone use it? That’s the most crucial question.
Step 4. Describe the Limitations in Your Discussion Section
Discussion section of a research paper isn’t limitless. What does that mean? Essentially, it means that you also have to discuss any limitations of your study. Maybe you had some methodological inconsistencies. Possibly, there are no particular theories or not enough information for you to be entirely confident in one’s conclusions. You might say that an available source of literature you have studied does not focus on one’s issue. That’s why one’s main limitation is theoretical. However, keep in mind that your limitations must possess a certain degree of relevancy. You can just say that you haven’t found enough books. Your information must be truthful to research.
Step 5. Conclude Your Discussion Section With Recommendations
Your last step when you write a discussion section in a paper is its conclusion, like in any other academic work. Writer’s conclusion must be as strong as their starting point of the overall work. Check out our brief list of things to know about the conclusion in research paper :
- It must present its scientific relevance and importance of your work.
- It should include different implications of your research.
- It should not, however, discuss anything new or things that you have not mentioned before.
- Leave no open questions and carefully complete the work without them.
Discussion Section of a Research Paper Example
All the best example discussion sections of a research paper will be written according to our brief guide. Don’t forget that you need to state your findings and underline the importance of your work. An undoubtedly big part of one’s discussion will definitely be answering and explaining the research questions. In other words, you’ll already have all the knowledge you have so carefully gathered. Our last step for you is to recollect and wrap up your paper. But we’re sure you’ll succeed!
How to Write a Discussion Section: Final Thoughts
Today we have covered how to write a discussion section. That was quite a brief journey, wasn’t it? Just to remind you to focus on these things:
- Importance of your study.
- Summary of the information you have gathered.
- Main findings and conclusions.
- Answers to all research questions without an open end.
- Correlation between literature review and your results.
But, wait, this guide is not the only thing we can do. Looking for how to write an abstract for a research paper for example? We have such a blog and much more on our platform.
Our academic writing service is just a click away. We are proud to say that our writers are professionals in their fields. Buy a research paper and our experts can provide prompt solutions without compromising the quality.
Discussion Section of a Research Paper: Frequently Asked Questions
1. how long should the discussion section of a research paper be.
Our discussion section of a research paper should not be longer than other sections. So try to keep it short but as informative as possible. It usually contains around 6-7 paragraphs in length. It is enough to briefly summarize all the important data and not to drag it.
2. What's the difference between the discussion and the results?
The difference between discussion and results is very simple and easy to understand. The results only report your main findings. You stated what you have found and how you have done that. In contrast, one’s discussion mentions your findings and explains how they relate to other literature, research questions, and one’s hypothesis. Therefore, it is not only a report but an efficient as well as proper explanation.
3. What's the difference between a discussion and a conclusion?
The difference between discussion and conclusion is also quite easy. Conclusion is a brief summary of all the findings and results. Still, our favorite discussion section interprets and explains your main results. It is an important but more lengthy and wordy part. Besides, it uses extra literature for references.
4. What is the purpose of the discussion section?
The primary purpose of a discussion section is to interpret and describe all your interesting findings. Therefore, you should state what you have learned, whether your hypothesis was correct and how your results can be explained using other sources. If this section is clear to readers, our congratulations as you have succeeded.

Joe Eckel is an expert on Dissertations writing. He makes sure that each student gets precious insights on composing A-grade academic writing.
You may also like

Want to create or adapt books like this? Learn more about how Pressbooks supports open publishing practices.
Results & Discussion
Characteristics of results & discussion.
- Results section contains data collected by scientists from experiments that they conducted.
- Data can be measurements, numbers, descriptions and/or observations.
- Scientific data is typically described using graphs, tables, figures, diagrams, maps, charts, photographs and/or equations.
- Discussion section provides an interpretation of the data, especially in context to previously published research.
The Results and Discussion sections can be written as separate sections (as shown in Fig. 2 ), but are often combined in a poster into one section called Results and Discussion. This is done in order to (1) save precious space on a poster for the many pieces of information that a scientist would like to tell their audience and (2) by combining the two sections, it becomes easier for the audience to understand the significance of the research. Combining the Results section and Discussion section in a poster is different for what is typically done for a scientific journal article. In most journal articles, the Results section is separated from the Discussion section. Journal articles are different from posters in that a scientist is not standing next to their journal article explaining it to a reader. Therefore, in a journal article, an author needs to provide more detailed information so that the reader can understand the research independently. Separating the Results section and Discussion section allows an author the space necessary to write a lengthier description of the research. Journal articles typically contain more text and more content (e.g., figures, tables) than posters.
The Results and Discussion section should contain data, typically in the form of a graph, histogram, chart, image, color-coded map or table ( Figs. 1 & 4 ). Very often data means numbers that scientists collect from making measurements. These data are typically presented to an audience in the form of graphs and charts to show a reader how these numbers change over time, space or experimental conditions ( Fig. 7 ). Numbers can increase, decrease or stay the same and a graph, or another type of figure, can be effectively used to convey this information to a reader in a visual format ( Fig. 7 ).
Figure 7. Example of a Graph
An audience will be attracted to a poster because of its figures and so it is very important for the author to pay particular attention to the creation, design and placement of the figures in a poster ( Figs. 1 & 4 ). A good figure is one that is informative, easy to comprehend and allows the reader to understand the significance of the data and experiment. Very often an author will use color to draw attention to a figure.
The Discussion section should state the importance of the research that is presented in the poster. It should provide an interpretation of the results, especially in context to previously published research. It may propose future experiments that need to be conducted as a result of the research presented in the poster. It should clearly illustrate the significance of the research with regards to new knowledge, understanding and/or discoveries that were made as part of the research.
Scientific Posters: A Learner's Guide Copyright © 2020 by Ella Weaver; Kylienne A. Shaul; Henry Griffy; and Brian H. Lower is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License , except where otherwise noted.
Share This Book
- Open access
- Published: 27 December 2022
Global patterns of diversity and metabolism of microbial communities in deep-sea hydrothermal vent deposits
- Zhichao Zhou 1 na1 ,
- Emily St. John 2 na1 ,
- Karthik Anantharaman 1 &
- Anna-Louise Reysenbach 2
Microbiome volume 10 , Article number: 241 ( 2022 ) Cite this article
7289 Accesses
17 Citations
126 Altmetric
Metrics details
When deep-sea hydrothermal fluids mix with cold oxygenated fluids, minerals precipitate out of solution and form hydrothermal deposits. These actively venting deep-sea hydrothermal deposits support a rich diversity of thermophilic microorganisms which are involved in a range of carbon, sulfur, nitrogen, and hydrogen metabolisms. Global patterns of thermophilic microbial diversity in deep-sea hydrothermal ecosystems have illustrated the strong connectivity between geological processes and microbial colonization, but little is known about the genomic diversity and physiological potential of these novel taxa. Here we explore this genomic diversity in 42 metagenomes from four deep-sea hydrothermal vent fields and a deep-sea volcano collected from 2004 to 2018 and document their potential implications in biogeochemical cycles.
Our dataset represents 3635 metagenome-assembled genomes encompassing 511 novel and recently identified genera from deep-sea hydrothermal settings. Some of the novel bacterial (107) and archaeal genera (30) that were recently reported from the deep-sea Brothers volcano were also detected at the deep-sea hydrothermal vent fields, while 99 bacterial and 54 archaeal genera were endemic to the deep-sea Brothers volcano deposits. We report some of the first examples of medium- (≥ 50% complete, ≤ 10% contaminated) to high-quality (> 90% complete, < 5% contaminated) MAGs from phyla and families never previously identified, or poorly sampled, from deep-sea hydrothermal environments. We greatly expand the novel diversity of Thermoproteia, Patescibacteria (Candidate Phyla Radiation, CPR), and Chloroflexota found at deep-sea hydrothermal vents and identify a small sampling of two potentially novel phyla, designated JALSQH01 and JALWCF01. Metabolic pathway analysis of metagenomes provides insights into the prevalent carbon, nitrogen, sulfur, and hydrogen metabolic processes across all sites and illustrates sulfur and nitrogen metabolic “handoffs” in community interactions. We confirm that Campylobacteria and Gammaproteobacteria occupy similar ecological guilds but their prevalence in a particular site is driven by shifts in the geochemical environment.
Our study of globally distributed hydrothermal vent deposits provides a significant expansion of microbial genomic diversity associated with hydrothermal vent deposits and highlights the metabolic adaptation of taxonomic guilds. Collectively, our results illustrate the importance of comparative biodiversity studies in establishing patterns of shared phylogenetic diversity and physiological ecology, while providing many targets for enrichment and cultivation of novel and endemic taxa.
Video Abstract
Introduction
Actively venting deep-sea hydrothermal deposits at oceanic spreading centers and arc volcanoes support a high diversity of thermophilic microorganisms. Many of these microbes acquire metabolic energy from chemical disequilibria created by the mixing of reduced high-temperature endmember hydrothermal fluids with cold oxygenated seawater. Community analysis of deposits using the 16S rRNA gene has revealed a rich diversity of novel archaeal and bacterial taxa [ 1 , 2 , 3 , 4 ] where the community composition is strongly influenced by the abundance of redox reactive species in high-temperature vent fluids (e.g., [ 5 , 6 , 7 ]). The variations in the composition of endmember fluids, and in turn the microbial community composition at different vent fields, reflect the temperature and pressure of fluid-rock interaction, in addition to substrate composition and entrainment of magmatic volatiles. For example, along the Mid-Atlantic Ridge, methanogens are associated with deposits from H 2 -rich vents at Rainbow and are absent in H 2 -poor vents at Lucky Strike [ 3 ]. At the Eastern Lau Spreading Center (ELSC), similar to other back-arc basins, the hydrothermal fluids are generally quite variable depending on differences in inputs of acidic magmatic volatiles, contributions from the subducting slab, and proximity of island arc volcanoes. Such geochemical differences are imprinted in the diversity of microbial communities [ 3 , 4 ]. Similar complex community structure dynamics have also been recently reported for the communities of the submarine Brothers volcano on the Kermadec Arc [ 8 ].
While such global patterns of high-temperature microbial diversity in deep-sea hydrothermal systems have demonstrated geological drivers of microbial colonization, little is known about the genomic diversity and physiological potential of the many reported novel taxa. While a few metagenomic studies of hydrothermal fluids and sediments have provided a much greater understanding of the functional potential of these communities (e.g., [ 7 , 9 , 10 , 11 , 12 , 13 ]), the metagenomic analysis of deposits has been limited to a small number of samples (e.g., [ 14 , 15 , 16 ]). One exception is the study of about 16 deep-sea hydrothermal deposits from Brothers volcano, which resulted in 701 medium- and high-quality metagenome-assembled genomes (MAGs) [ 8 ]. Further, this study demonstrated that there were functionally distinct high-temperature communities associated with the volcano that could be explained through an understanding of the geological history and subsurface hydrologic regime of the volcano.
Here, we expand on the Brothers volcano study by exploring the genomic and functional diversity of hydrothermal deposits collected from deep-sea vents in the Pacific and Atlantic oceans. We greatly increase the number of novel high-quality assembled genomes from deep-sea vents, many of which are endemic to vents and do not have any representatives in culture yet. We also show that known important biogeochemical cycles in hydrothermal ecosystems are accomplished by the coordination of several taxa as metabolic handoffs, where in some cases different taxa accomplish similar functions in different environments, potentially providing functional redundancy in fluctuating conditions.
Results and discussion
Patterns of metagenomic diversity in deep-sea hydrothermal deposits.
We sequenced 42 metagenomes from 40 samples (38 hydrothermal vent deposit samples and two diffuse flow fluids) collected at deep-sea hydrothermal vents and a deep-sea volcano. These represent one of the largest global collections of metagenomes from such samples (Fig. S 1 , S 2 ). This study spans vent deposit collections from 2004 to 2018, from deep-sea hydrothermal vent fields in the north Atlantic (Mid-Atlantic Ridge, MAR), east and southwest Pacific (East Pacific Rise, EPR; Eastern Lau Spreading Center, ELSC), a sedimented hydrothermal system (Guaymas Basin, GB), and a deep-sea volcano (Brothers volcano, BV) (Table S 1 ).
In this study, de novo assembly of sequencing data and subsequent genome binning and curation (see the “ Methods ” section for details) resulted in 2983 bacterial and 652 archaeal draft metagenome-assembled genomes (MAGs with ≥ 50% completeness, Table S 2 ). Of these, ~ 21% were > 90% complete, with < 5% contamination, and ~ 36% contained a 16S rRNA gene fragment. The MAGs were initially characterized phylogenetically using the Genome Taxonomy Database Toolkit (GTDB-Tk) (Figs. 1 , 2 , and 3 , Data S 1 , S 2 , S 3 , S 4 , S 5 ) [ 17 ]. MAGs that could not be assigned to a known genus by GTDB-Tk were assigned to new genera using AAI with the recommended cutoffs in Konstantinidis et al. [ 18 ] (Table S 3 A, B). Shared phyla between most of the hydrothermal deposits (excluding samples from the highly acidic Brothers volcano sites, and the diffuse flow fluids) included the Halobacteriota (e.g., Archaeoglobaceae), Methanobacteriota (e.g., Thermococcaceae), Thermoproteota (e.g., Acidilobaceae, Pyrodictiaceae), Acidobacteriota, Aquificota (e.g., Aquificaceae), Bacteroidota (e.g., Flavobacteriaceae), Campylobacterota (e.g., Sulfurimonadaceae, Nautiliaceae, Hippeaceae), Chloroflexota, Deinococcota (e.g., Marinithermaceae), Desulfobacterota (e.g., Dissulfuribacteraceae, Thermodesulfobacteriaceae), Proteobacteria (e.g., Alphaproteobacteria, Gammaproteobacteria), and the Patescibacteria (Table S 4 ). Many of these phyla have only a few representatives in isolated cultures and point to the importance of combining enrichment cultivation strategies with metagenomic approaches to obtain additional insights into the physiological ecology of these core lineages.
Maximum-likelihood phylogenomic tree of bacterial metagenome-assembled genomes, constructed using 120 bacterial marker genes in GTDB-Tk. Major taxonomic groups are highlighted, and the number of MAGs in each taxon is shown in parentheses. See Table S 2 for details. Bacterial lineages are shown at the phylum classification, except for the Proteobacteria which are split into their component classes. The inner ring displays quality (green: high quality, > 90% completion, < 5% contamination; purple: medium quality, ≥ 50% completion, ≤ 10% contamination), while the outer ring shows normalized read coverage up to 200x. The scale bar indicates 0.1 amino acid substitutions per site, and filled circles are shown for SH-like support values ≥ 80%. The tree was artificially rooted with the Patescibacteria using iTOL. The Newick format tree used to generate this figure is available in Data S 4 , and the formatted tree is available online at https://itol.embl.de/shared/alrlab
Maximum-likelihood phylogenomic reconstruction of deep-sea hydrothermal vent archaeal metagenome-assembled genomes generated in GTDB-Tk. The tree was generated with 122 archaeal marker genes. Taxa are shown at the phylum level, except for the Thermoproteota, Asgardarchaeota, Halobacteriota, and Methanobacteriota, shown at the class level. The number of MAGs in each highlighted taxon is shown in parentheses. See Table S 2 for details. Quality is shown on the inner ring (green: high quality, purple: medium quality, with one manually curated Nanoarchaeota MAG below the 50% completion threshold also displayed as medium quality), while the outer ring displays normalized read coverage up to 200x. SH-like support values ≥ 80% are indicated with filled circles, and the scale bar represents 0.1 amino acid substitutions per site. The tree was artificially rooted with the Iainarchaeota, Micrarchaeota, SpSt-1190, Undinarchaeota, Nanohaloarchaeota, EX4484-52, Aenigmarchaeota, Aenigmarchaeota_A, and Nanoarchaeota using iTOL. The tree used to create this figure is available in Newick format (Data S 5 ), and the formatted tree is publicly available on iTOL at https://itol.embl.de/shared/alrlab
Relative abundance of MAG phyla, based on normalized read coverage. The phyla shown comprise ≥ 10% of the MAG relative abundance in at least one metagenomic assembly. Read coverage was normalized to 100 M reads per sample, and coverage values for MAGs were summed and expressed as a percent. UC, Upper Cone; LC, Lower Cone, NWC-A, Northwest Caldera Wall A; NWC-B, Northwest Caldera Wall B and Upper Caldera Wall; DF, diffuse flow; VL, Vai Lili; RB, Rainbow; LS, Lucky Strike
While shared taxa differed in relative abundance and distribution, observable differences in community structure between vent fields were somewhat limited in this study due to small sample numbers from some of the vent fields (two samples apiece from EPR; Rainbow, MAR; Lucky Strike, MAR), and the overall lower read depth of samples from these sites and a few other samples (Fig. S 3 ). Therefore, obtaining statistically robust community structure patterns using MAG phylogenetic diversity for the entire dataset was not possible. However, Reysenbach et al. [ 8 ] did show that if metagenomic sequencing is deep, assembled MAG diversity tracks 16S rRNA amplicon diversity structure. Extrapolating to this study, the Brothers volcano MAG diversity patterns were retained and confirmed the amplicon observations from Reysenbach et al. [ 8 ] (Fig. S 4 ), and in turn tracked the ELSC MAG community diversity (Fig. 4 A, B). For example, sites at Brothers volcano that were hypothesized to have some magmatic inputs were predicted to be more similar in community structure to the sites along the ELSC with greater magmatic inputs, such as Mariner. Several of the samples from the more acidic Mariner vent field were more closely aligned in MAG diversity structure to those of the acidic solfataric Upper Cone sites at Brothers. The MAG data also demonstrated that the Guaymas samples were quite unique, which is not surprising, given that Guaymas Basin is a sediment-hosted system where the hydrothermal fluid geochemistry is quite different from other basalt- or andesitic-hosted hydrothermal systems (e.g., higher pH, high organics, high ammonia and methane) [ 19 , 20 ].
Non-metric multidimensional scaling (NMDS) plots showing taxonomic diversity of MAGs. Plots depict A all samples in this study and B a subset of the data, limited to locations with three or more samples. Plots were generated using Bray–Curtis matrices of the relative abundance of GTDB taxa, based on normalized read coverage of medium- and high-quality MAGs (Table S 4 ; set to 100 M reads and expressed as a percentage of MAG read coverage per sample). Points that are closer together in the plots represent a higher degree of similarity
Our dataset greatly broadens genomic diversity from deep-sea vents, by representing 511 novel and previously identified [ 8 ] genera, comprising 395 Bacteria and 116 Archaea. Notably, 52% (206) of these bacterial genera (Table S 3 A) and 72% (84) of archaeal genera (Table S 3 B) were found at Brothers volcano. Furthermore, 25% (99) of the recently identified bacterial genera and 47% (54) of the archaeal genera were unique to the Brothers volcano samples (Tables S 3 A, B), which further supports the understanding that this environment is a hotbed for novel microbial biodiversity, reflected in the volcano’s complex subsurface geology [ 8 ].
While many of these novel archaeal and bacterial genera were previously reported from Brothers volcano [ 8 ], we report them again here in the context of the new data of the four deep-sea hydrothermal vent environments and the new assemblies (1000 bp contig cutoff, used for Brothers volcano samples and ELSC 2015 samples) and iterative DAS Tool binning used for all our metagenomes. Our data support that of Reysenbach et al. [ 8 ], which used MetaBAT for assemblies (2000 bp contig cutoff) of the Brothers volcano metagenomes. Namely, we recovered approximately 202 novel bacterial genera and 83 new archaeal genera from Brothers volcano communities in Reysenbach et al. [ 8 ], well within the range detected in this analysis (viz. 206 and 84, respectively). In this study, using a lower contig cutoff allowed for the recovery of a much higher number of MAGs, but many are of lower quality with higher contig counts. For example, MAGs recovered in the Reysenbach et al. [ 8 ] study had an average of 254 contigs per MAG, with ~ 19% (135) of MAGs comprising 100 contigs or less. In contrast, only 7% (258) of MAGs in this current study had 100 contigs or less, and the average number of contigs per MAG was 511 (Table S 2 ). However, using the iterative binning approach provided advantages when resolving lineages of high microdiversity, such as in the Nautiliales, with the caveat of creating some MAGs with large collections of erroneous contigs that were poorly detected by CheckM, as they had very few associated marker genes (e.g., MAGs 4571-419_metabat1_scaf2bin.008, M10_maxbin2_scaf2bin.065; Fig. S 5 ). This points to the importance of carefully choosing assembly parameters depending on the ultimate goal of whether quality over quantity of MAGs is preferred for analyses of ecological patterns. Our data demonstrate, however, that overall patterns of MAG diversity are retained regardless of assembly techniques and parameters (Fig. S 4 ).
Furthermore, here we document some of the first examples of medium- to high-quality MAGs from phyla and classes never previously identified, or poorly sampled, from deep-sea hydrothermal environments. These include Thermoproteia, Patescibacteria (formerly Candidate Phyla Radiation, CPR), Chloroflexota, and a few MAGs representing two putative new bacterial phyla, JALSQH01 (3 MAGs) and JALWCF01 (13 MAGs) (Supplementary Discussion, Fig. S 6 , Table S 5 ). For example, with 249 MAGs belonging to the Thermoproteia (Table S 2 , Fig. S 7 ), we have significantly expanded the known diversity and genomes from this phylum. The importance of this group at deep-sea vents was first recognized through 16S rRNA amplicon studies, where the depth of sequencing highlighted that much of this novel thermophilic diversity had been overlooked (e.g., [3, 4]). Furthermore, it is now recognized that many members of this group have several introns in the 16S rRNA gene, which explains why they were missed in original clone library assessments and may be underestimated in amplicon sequencing [ 21 , 22 , 23 , 24 ]. For example, 24 MAGs were related to a recently described genus of the Thermoproteia, Zestosphaera (GTDB family NBVN01) [ 24 ]. This genus was first isolated from a hot spring in New Zealand but is clearly a common member of many deep-sea vent sites. Further, the discovery of a 16S rRNA gene related to Caldisphaera at deep-sea vents [ 25 ], previously only detected in terrestrial acidic solfataras, led to the isolation of related Thermoplasmata— Aciduliprofundum boonei —but the Caldisphaera escaped cultivation. Here we report several high-quality MAGs related to this genus (M2_metabat2_scaf2bin.319, 131-447_metabat1_scaf2bin.050, M1_metabat1_scaf2bin.025, S016_metabat2_scaf2bin.003). Additionally, we also recovered a genome from the Gearchaeales (S146_metabat1_scaf2bin.098), first discovered in iron-rich acidic mats in Yellowstone National Park [ 26 ], and members of the poorly sampled Ignicoccaceae, Ignisphaeraceae, and Thermofilaceae. While we identified several genomes from recently discovered archaeal lineages including the Micrarchaeota, Iainarchaeota, and Asgardarchaeota, we also recovered 15 MAGs belonging to the Korarchaeia, 14 of which comprise two putative novel genera, and one which is closely related to a MAG previously recovered from sediment in Guaymas Basin (Genbank accession DRBY00000000.1) [ 27 , 28 ]. Additionally, we recovered four MAGs from the Caldarchaeales that span two novel genera, one of which was recently proposed as Candidatus Benthortus lauensis [ 29 ] using a MAG generated from a previous assembly of the T2 metagenome (T2_175; Genbank accession JAHSRM000000000.1). MAGs belonging to this genus were identified at both Tui Malila, ELSC, and Brothers volcano (T2_metabat2_scaf2bin.284, S140_maxbin2_scaf2bin.281, S141_maxbin2_scaf2bin.262) with the Tui Malila MAG nearly identical (99.7% AAI similarity) to the described Cand . B. lauensis T2_175 MAG.
While within the Bacteria, the Gammaproteobacteria and Campylobacterota were by far the most highly represented bacterial genomes, there were other lineages for which we have very little if any data or cultures from deep-sea hydrothermal systems (Fig. 3 , Fig. S 7 ). Two such groups are the Patescibacteria and Chloroflexota, with 154 and 194 MAGs respectively.
Patescibacteria and Chloroflexota are diverse and abundant members of deep-sea hydrothermal vent deposits
The Patescibacteria/Candidate Phyla Radiation (CPR) encompasses a phylogenetically diverse branch within the bacterial tree of life that is poorly understood and rarely documented in deep-sea hydrothermal systems. Originally, the CPR was proposed to include several phylum-level lineages [ 30 ], but the entire group was later reclassified by GTDB as a single phylum, Patescibacteria [ 31 ]. Members of the Patescibacteria have been well-characterized in terrestrial soils, sediments, and groundwater [ 32 , 33 , 34 , 35 , 36 , 37 ], and in the mammalian oral cavity [ 38 , 39 , 40 ]. Several 16S rRNA gene and metagenomic studies have also identified members of the Patescibacteria from deep-sea vents, including EPR, MAR, ELSC, and Guaymas Basin [ 3 , 4 , 12 , 15 , 41 , 42 , 43 ], from Suiyo Seamount [ 44 ], and the Santorini submarine volcano [ 45 ], further supporting the widespread distribution of this metabolically diverse phylum.
Our study adds 56 novel genera based on AAI and GTDB classifications to the Patescibacteria phylum. These include large clades within the Gracilibacteria (10 new genera), representatives within the Microgenomatia (9 novel genera), Dojkabacteria (10 new genera), and several clades in the Paceibacteria (13 new genera) (Fig. 5 A, B , Fig. S 8 ). The Gracilibacteria and Paceibacteria were overall the most prevalent lineages of Patescibacteria in the samples but had contrasting distributions across vents (Fig. 5 B). In general, when the Gracilibacteria were prevalent, the Paceibacteria appeared to be a minor component or not present, and vice versa. In particular, the Gracilibacteria MAGs were often associated with the acidic sites such as the Upper Cone at Brothers volcano (S011, S147), and the Mariner vent fields, and in the early colonization experiment from Guaymas Basin (Supplementary Discussion). This may suggest that Gracilibacteria function as early colonizers and are associated with turbulent ephemeral environments as observed previously in oil seeps [ 46 ]. Continued investigation into the ecology, evolution, and host association patterns of these groups, however, may shed more light on these distribution differences.
Phylogenomic placement and relative abundance of Patescibacteria MAGs, displayed at the class rank. A Blue clades in the maximum-likelihood phylogenomic tree contain MAGs from this study, with the number of MAGs shown in parentheses. The scale bar shows 0.5 substitutions per amino acid, and filled circles indicate SH-like support (≥ 80%). B Relative abundance of Patescibacteria MAGs was calculated using normalized read coverage for MAGs in each assembly (set to 100 M reads and expressed as a percentage of MAG read coverage per sample)
Consistent with previous studies [ 30 , 34 ], many of the recovered Patescibacteria MAGs had very small genomes (often ~ 1 MB or smaller; Table S 2 ) with highly reduced metabolic potential, often lacking detectable genes for synthesis of fatty acids, nucleotides, and most amino acids (Table S 6 ). Gene patterns also suggested that many of the organisms are obligate anaerobes, lacking aerobic respiration, and that they likely form symbiotic or parasitic associations with other microbes, as has been shown for Patescibacteria cultivated thus far from the Absconditabacterales and Saccharibacteria [ 39 , 40 , 47 , 48 ].
We recovered several MAGs from Mariner, Guaymas Basin, and Brothers volcano that were related to the parasitic Cand . Vampirococcus lugosii [ 47 ] and Cand . Absconditicoccus praedator [ 48 ]. In order to explore if our MAGs had any hints of a parasitic lifestyle, we searched for some of the large putative cell-surface proteins identified in the genomes of Cand . V. lugosii [ 47 ] and Cand . A. praedator [ 48 ]. Using a local BlastP of nine of the longest genes found in Cand . V. lugosii, we recovered high-confidence homologs ( E -value = 0) for alpha-2 macroglobulin genes in several MAGs from the Abscontitabacterales (based on search of Cand . V. lugosii protein MBS8121711.1), which may be involved in protecting parasites against host defense proteases [ 47 ]. We also recovered homologs for PKD-repeat containing proteins (MBS8122536.1; E -value = 0), which are likely involved in protein–protein interactions [ 47 ]. Previous analysis of Cand . V. lugosii found these giant proteins are likely membrane-localized, suggesting they may potentially play a role in host/symbiont interactions. Additionally, we identified these long proteins from Cand . V. lugosii elsewhere in the Gracilibacteria MAGs. For example, putative homologs of the PKD repeat containing protein (MBS8122536.1), a hypothetical protein (MBS8121701.1), and the alpha-2 macroglobulin (MBS8121711.1) were identified in multiple other orders of the class Gracilibacteria ( E -value ≤ 1E − 25). The alpha-2 macroglobulin was also identified in the very distantly related Paceibacteria, and a single putative homolog of the alpha-2 macroglobulin was found in a MAG belonging to the class WWE3 (134-614_metabat1_scaf2bin.084; E -value ≤ 1E − 24).
While the Patescibacteria likely rely on symbiotic or parasitic relationships, members of the Chloroflexota phylum are diverse and metabolically flexible organisms, capable of thriving in a wide variety of geochemical niches. Chloroflexota are abundant and widely distributed in a variety of environments, including terrestrial soils, sediments and groundwater, freshwater, pelagic oceans, and the marine subseafloor and sediments [ 49 , 50 , 51 , 52 , 53 , 54 , 55 ], and hydrothermal settings such as Guaymas Basin [ 11 ] and Brothers submarine volcano [ 8 ]. Genomic evidence suggests that Chloroflexota are associated with important metabolisms in the carbon cycle, including fermentation, carbon fixation, acetogenesis, and the utilization of sugars, polymers, fatty acids, organic acids, and other organic carbon compounds [ 50 , 51 , 54 ].
Here we add to the growing evidence that the Chloroflexota are diverse and metabolically versatile members of deep-sea hydrothermal vent communities. We recovered a total of 194 Chloroflexota MAGs spanning 12 orders (GTDB taxonomy), which included 22 novel genera. Of these novel genera, 14 were identified at Brothers volcano and 6 were unique to the Brothers volcano samples (Table S 3 A). Based on read coverage, Chloroflexota MAGs were in high relative abundance (≥ 7%) in several samples from the ELSC, namely, from Tui Malila and ABE, and in one NW Caldera Wall sample from Brothers volcano (Table S 4 ). To further explore the metabolic potential of Chloroflexota in hydrothermal vent communities, we focused our analyses on ≥ 80%-completeness MAGs (≥ 80% completeness, n = 58) distributed in 6 orders: Caldilineales, Promineofilales, Anaerolineales, Ardenticatenales, B4-G1, and SBR1031 (Fig. 6 , Table S 7 A).

Phylogenetic tree of 58 ≥ 80%-completeness Chloroflexota MAGs with predicted functional capabilities. Nodes with ultrafast bootstrap support values ≥ 90% are shown with filled circles, and the scale bar shows 0.2 substitutions per site. One genome from the GTDB r202 database (GTDB accession GB_GCA_007123655.1) was used to re-root the tree. Hydrothermal vent fields: Brothers volcano (green), Eastern Lau Spreading Center (blue), East Pacific Rise (orange), Mid Atlantic Ridge (yellow)
The majority (≥ 75%) of the ≥ 80%-completeness Chloroflexota MAGs encoded marker genes involved in several processes previously associated with the Chloroflexota (Table S 7 B), including fatty acid degradation [ 50 , 55 ], formate oxidation [ 56 ], aerobic CO oxidation [ 57 ], and selenate reduction [ 53 ]. Except for the Anaerolineales, over 66% of the MAGs in the other five orders had the capacity for degradation of aromatic compounds, as previously reported for Chloroflexota from the marine subsurface [ 51 ]. While some MAGs had the potential for substrate-level phosphorylation through acetate formation, most of the MAGs contained pathways for oxidative phosphorylation and oxygen metabolism [ 50 , 51 ]. The Wood–Ljungdahl pathway, the CBB cycle based on a Form I Rubisco, and the reverse TCA cycle were detected in some of the MAGs [ 50 , 51 ]. Soluble methane monooxygenase genes, a metabolic potential recently also detected in a Chloroflexota MAG from the arctic [ 58 ], were identified in a total of eight of our MAGs from the orders Caldilineales, Anaerolineales, and Ardenticatenales.
Although the primary metabolic potential of the hydrothermal vent-associated Chloroflexota was in carbon cycling, we did, however, observe minor evidence for their roles in nitrogen and sulfur cycling (Fig. 6 , Table S 7 ). About 22% of the MAGs (with ≥ 80% completeness) encoded capacities for sulfide oxidation, as previously reported for members of this group, e.g., Chloroflexus spp. [ 59 , 60 ]. The potential to disproportionate thiosulfate was also observed in a few MAGs. Further, thermophilic Chloroflexota grown in an enrichment culture from Yellowstone National Park were shown to oxidize nitrite. A few of our MAGs encoded genes involved in nitrite oxidation [ 61 ], while a larger proportion of the MAGs encoded genes for nitrite or nitric oxide reduction. None of the MAGs encoded complete pathways for entire sulfur oxidation or denitrification, suggesting that Chloroflexota in these environments may be associated with metabolic handoffs involving other community members (see below).
Metabolic and functional diversity in deep-sea hydrothermal vent deposits
In order to explore the metabolic and functional diversity associated with our MAGs, we utilized functional assignment results in tandem with the corresponding MAG relative abundance (Table S 8 ). In general, genes involved in carbon, nitrogen, sulfur, and hydrogen metabolism were prevalent and shared across all hydrothermal systems in this study (Figs. 7 and 8 ). While heterotrophy, autotrophy, and mixotrophy potential were identified in all samples, 47.1% of the MAGs (by count) exhibited potential for carbon fixation. Marker genes associated with five different carbon fixation pathways were identified in the MAGs, namely, the Calvin-Benson-Bassham (CBB) cycle (form I or form II Rubisco), the 3-hydroxypropionate/4-hydroxybutyrate cycle, the dicarboxylate/4-hydroxybutyrate cycle, the reverse TCA cycle, and the Wood–Ljungdahl pathway (Figs. 7 and 8 ). Marker gene presence also suggested the potential for widespread heterotrophic metabolism of peptides, polysaccharides, nucleotides, and lipids, and fermentation via acetogenesis (Figs. 7 and 8 ).

Core metabolic gene presence across phylogenetic clusters in deep-sea hydrothermal vent deposits. The number of MAGs in each clade is shown in parentheses, and MAGs belonging to unclassified lineages or falling outside their corresponding phylogenetic cluster due to unstable tree topology are shown without names. In instances where a phylum was not recovered as a monophyletic lineage within the tree (e.g., Iainarchaeia), MAG count and gene distribution for the entire phylum is only shown on one of the branches. Unless otherwise indicated, archaeal clades are shown at the class level, while bacterial clades are shown at the phylum level. Nodes with ultrafast bootstrap support ≥ 90% are shown with filled circles, and scale bars indicating 0.2 amino acid substitutions per site are provided for both archaeal and bacterial trees. Detailed metabolic gene presence information can be found in Table S 9
Heatmap displaying the metabolic potential for each metagenome. Within each metagenomic dataset, functional abundance values were calculated as described in the methods. Functional abundances were then log-transformed, with abundance values equal to zero replaced by 10 −3 to avoid negative infinite values
Genes involved in nitrogen fixation, denitrification, and nitrite oxidation were identified across the different hydrothermal sites, yet the potential for anaerobic or aerobic ammonia oxidation was rarely detected (Fig. 8 ). The absence of ammonia oxidation is not totally surprising, since ammonia is in very low to undetectable concentrations in deep-sea hydrothermal fluids, with the exception of sediment-hosted hydrothermal areas like at Guaymas Basin [ 19 , 20 ]. In these sedimented hydrothermal systems, aerobic and anaerobic ammonia oxidation are key processes within the sediments and hydrothermal plumes [ 62 , 63 , 64 , 65 ], but they may not be as important in the hydrothermal deposits. Our data also expands the importance of nitrogen fixation from the first detection at deep-sea vents in Methanocaldococcus [ 66 ] to a greater diversity of hydrothermal Bacteria and Archaea.
Given the importance of sulfur cycling in deep-sea hydrothermal systems [ 67 , 68 , 69 ], it is not surprising that genes associated with elemental sulfur, sulfide, and thiosulfate oxidation; sulfate reduction; and thiosulfate disproportionation were widely distributed in MAGs from different hydrothermal samples and were associated with diverse taxonomic guilds (Figs. 7 and 8 ). Based on metabolic gene distribution statistics (Table S 9 ), the potential for sulfur oxidation was identified in 16% of the MAGs (577), primarily in members of the Alphaproteobacteria and Gammaproteobacteria. Genes associated with sulfide oxidation were identified in 34% of the MAGs (1216), including members of the Bacteroidia, Campylobacteria, Alphaproteobacteria, and Gammaproteobacteria. Thiosulfate oxidation genes were detected in 23% of the MAGs (836), largely comprised of the Campylobacteria, Alphaproteobacteria, and Gammaproteobacteria, while 14% of the MAGs (522) encoded genes for thiosulfate disproportionation, including the classes Bacteroidia and Campylobacteria and the phylum Desulfobacterota. The potential for dissimilatory sulfite reduction was identified in 6% of the MAGs (220) distributed across ten bacterial and archaeal phyla, namely Halobacteriota (class Archaeoglobi), Bacteroidota (class Kapabacteria), Campylobacterota (class Campylobacterales), Zixibacteria, Gemmatimonadota, Acidobacteriota, Nitrospirota, Desulfobacterota, Desulfobacterota_F, and Myxococcota.
Hydrogen is highly variable in hydrothermal fluids, with some of the highest concentrations in geothermal systems hosted by ultramafic rocks, such as the Rainbow hydrothermal vent field [ 3 ], or in sediment-hosted regions like Guaymas basin [ 70 ]. In these systems, methanogens and sulfate reducers are prevalent hydrogen consumers [ 3 , 71 , 72 , 73 , 74 ], although a wide variety of other heterotrophs and autotrophs can also derive energy from hydrogen oxidation [ 72 ]. Hydrogenase enzymes are responsible for mediating hydrogen oxidation in microbial populations but are also involved in a variety of other functions, including hydrogen evolution, electron bifurcation, and hydrogen sensing [ 75 ]. Approximately 27% of the MAGs in this study (974) encoded for at least one hydrogenase gene for hydrogen oxidation, and the MAGs were predominantly associated with the classes Campylobacteria, Bacteroidia, Gammaproteobacteria, and the phylum Desulfobacterota (Figs. 7 and 8 , Table S 9 ). In several cases (132 MAGs), hydrogenase genes co-occurred with genes involved in the oxidation of reduced sulfur species (sulfide, elemental sulfur, sulfite, or thiosulfate). This is not surprising, given that the capability to oxidize both sulfur and hydrogen has been shown in multiple isolates, including members of the Campylobacteria [ 76 , 77 , 78 ] and Aquificae (e.g., [ 79 , 80 ]).
Metabolic handoffs are a central feature of community interactions in deep-sea hydrothermal vent deposits
The microbial communities at deep-sea hydrothermal vents are shaped by a wide variety of complex interactions, including symbiosis, syntrophy, commensalism, cross-feeding, and metabolic handoffs [ 11 , 12 , 81 , 82 , 83 ]. While many of the MAGs encode genes associated with different biogeochemical cycles, as expected, the genes for a complex functional pathway often were not localized in a single MAG, but instead distributed across several MAGs. This is likened to “metabolic handoffs” where the interaction between different organisms produces pathway intermediates, enabling community members to perform downstream reactions in the metabolic pathway. For example, metagenomic analysis of a subsurface aquifer environment suggested that metabolic handoffs are commonly utilized in key biogeochemical pathways such as sulfide oxidation and denitrification [ 37 ]. Genes for sulfide oxidation were identified in all the deep-sea hydrothermal vent sites in this study, but few MAGs encoded genes for the entire three-step pathway. A much larger proportion of the MAGs, however, contained genes for a single step in sulfur oxidation (Fig. 9 ), consistent with a metabolic handoff scenario. Similar patterns were also observed for sulfate reduction and denitrification (Fig. 9 ). Additionally, the genes for individual steps in sulfide oxidation were often found coupled with at least one gene from the denitrification pathway, which may increase the thermodynamic favorability of both pathways. Furthermore, one or more denitrification genes co-occurred with sulfide oxidation genes in 1113 MAGs, with elemental sulfur oxidation genes in 485 MAGs and with sulfite oxidation genes in 1025 MAGs (Table S 9 ). We recognize that some of these observations may be attributed to the incompleteness of the MAGs; however, our observations are in line with similar findings from other environments such as the terrestrial subsurface [ 37 ].
Bar plots showing the sequential steps of sulfur oxidation, denitrification, and sulfate reduction. Bar height indicates the percent relative abundance of MAGs in each metagenome with genes for a particular function(s), averaged across hydrothermal vent sites
Conserved microbial functions are mediated by different taxa at different hydrothermal vent systems
Previous analyses of deep-sea hydrothermal environments and global oceans have pointed to widespread functional redundancy in microbial communities [ 8 , 12 , 84 , 85 ], with similar metabolic potential identified across taxonomically diverse samples. For example, a study of Guaymas Basin metagenome-assembled genomes suggested that many functional genes could be identified across multiple distinct taxa [ 12 ]. In our study, members of the Campylobacteria and Gammaproteobacteria were present in almost all samples, yet showed contrasting patterns of abundance (Fig. 10 ). These lineages can perform several of the same functional processes including oxidation of reduced sulfur species [ 86 ], denitrification [ 87 , 88 , 89 ], and carbon fixation [ 90 , 91 , 92 , 93 ]. This can be partially explained by ecophysiological and growth differences between the groups, which are selected for by the different geochemical profiles at the various vent sites. For example, studies have suggested that Campylobacteria tend to favor higher sulfide conditions but have a broader range of oxygen tolerance than the Gammaproteobacteria, while Gammaproteobacteria tend to inhabit a narrower range of higher oxygen and lower sulfide [ 16 , 86 , 90 , 94 ]. It is therefore not surprising that the Campylobacteria were more prevalent at several of the acidic and more turbulent sites, such as at the Upper Cone, Brothers volcano, and in early colonized samples from a thermocouple array at Guaymas Basin (Table S 4 , Supplementary Discussion). Patwardhan et al. [ 95 ] also showed that Campylobacteria were early colonizers of shallow marine vents followed by Gammaproteobacteria, and their differential colonization could be linked to sulfide, oxygen, and temporal differences.
Comparative taxonomic and functional gene abundance of the Campylobacteria and Gammaproteobacteria. NMDS plots were generated using a Bray–Curtis matrix of relative MAG abundance, based on GTDB-assigned taxonomy at the class level. Plots are shown for A all sample sites, and for all sample sites with bubbles proportional to the relative abundance of B Gammaproteobacteria and C Campylobacteria. D Comparative functional distribution is also shown for the Gammaproteobacteria and Campylobacteria for the 26 samples that had a summed relative abundance of both Gammaproteobacteria and Campylobacteria of ≥ 30%. The 22 functions depicted were selected as the Gammaproteobacteria and Campylobacteria accounted for an average of ≥ 20% of the total abundance for each function across the metagenomes
The covariation of the Campylobacteria and Gammaproteobacteria in our data also coincided with genes for key functional processes associated with these taxa (Fig. 10 ). Thus, the overall ecological function contributed by the Campylobacteria and Gammaproteobacteria to the community at all sites was similar, but carried out by either one, viz., same guild different taxa. For example, relative gene abundance of individual functions tracked the relative abundance of Campylobacteria and Gammaproteobacteria for 15 of 22 broadly distributed functions, including heterotrophy associated with various organic carbon compounds, respiration of oxygen and nitrogen compounds, and oxidation of reduced sulfur compounds. However, genes for some functions were exclusively represented by either group (Fig. 10 , Table S 10 ). For example, marker genes for formaldehyde oxidation, urea utilization, and elemental sulfur oxidation were found in the Gammaproteobacteria but were hardly detected in Campylobacteria, while genes associated with thiosulfate disproportionation were attributed almost exclusively to Campylobacteria (Fig. 10 , Table S 10 ). In some cases, metabolic analysis also suggested that both Campylobacteria and Gammaproteobacteria had similar metabolic capabilities but encoded different pathways for the same functions. For example, consistent with a previously observed but non-ubiquitous trend [ 90 , 91 , 92 , 93 ], Campylobacteria mostly encoded genes for the rTCA cycle while the Gammaproteobacteria encoded genes for the CBB cycle. Both taxa also showed the potential for nitrite reduction to ammonia, with more nrfADH genes identified in the Campylobacteria and nirBD only found in the Gammaproteobacteria.
Conclusions
From a comparative metagenomic analysis of 38 deep-sea hydrothermal deposits from multiple globally distributed sites, we provide insights into the shared vent-specific lineages and greatly expand the genomic representation of core taxa that have very few, if any, examples in cultivation. Furthermore, we document many novel high-quality assembled genomes that were originally only identified from deep-sea vents as 16S rRNA genes. This study sheds light on the metabolic potential and physiological ecology of such taxa. We show that overall, the different communities share similar functions, but differences in the environmental geochemistry between sites select distinct taxonomic guilds. Further, metabolic handoffs in communities provide functional interdependency between populations achieving efficient energy and substrate transformation, while functional redundancy confers higher ecosystem resiliency to perturbations and geochemical fluctuations. In summary, this study provides an integrated view of the genomic diversity and potential functional interactions within high-temperature deep-sea hydrothermal deposits and has implications on their biogeochemical significance in mediating energy and substrate transformations in hydrothermal environments.
Sample collection, DNA extraction, and sequencing
High-temperature, actively venting deep-sea hydrothermal deposits, a diffuse flow sample, and a water sample were collected from Brothers volcano (2018), the Eastern Lau Spreading Center (2005 and 2015), Guaymas Basin (2009), the Mid-Atlantic Ridge (2008), and the East Pacific Rise (2004 and 2006) as previously described (Flores et al., 2012a, Reysenbach et al., 2020). Expedition details, including identification numbers, research vessels, and submersibles utilized for sampling, are described in Table S 1 . Samples were processed [ 4 ] and DNA extraction was performed as previously described [ 4 , 8 , 25 , 96 ].
Thermocouple array from Guaymas Basin
The thermocouple array experimental setup from Guaymas Basin in 2009 is described in Teske et al. [ 20 ].
Metagenomic assembling and binning
Reads from Brothers volcano and ELSC (2015) were quality-filtered using FastQC v.0.11.8 ( https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ ) and de novo assembled using metaSPAdes v.3.12.0 [ 97 ] with the settings “-k 21,33,55,77,99,127 -m 400 –meta”. Reads from ELSC (2005), MAR, EPR, and Guaymas Basin were assembled by the Department of Energy, Joint Genome Institute (JGI) using metaSPAdes v.3.11.1 with the settings “-k 33,55,77,99,127 –only-assembler –meta”. Individual assemblies were generated for each metagenomic dataset. MetaWRAP v.1.2.2 [ 98 ] was used to generate metagenome-assembled genomes (MAGs) from each assembly with the settings “–metabat2 –metabat1 –maxbin2”. DAS Tool v.1.0 [ 99 ] was then applied to screen the three sets of MAGs generated by MetaWRAP, resulting in consensus MAGs with a minimum scaffold length of 1000 bp.
Metagenome-assembled genome curation and quality assessment
CheckM v.1.0.7 [ 100 ] was used to assess MAG quality and screen for the presence of 16S rRNA genes. Erroneous SSU genes were then removed using RefineM v.0.0.20 [ 101 ], which was also used to identify and remove outlier scaffolds with abnormal coverage, tetranucleotide signals, and GC patterns from highly contaminated MAGs. GTDB-Tk v.1.5.0, data release 202 [ 17 ], was used to assign taxonomy to each MAG with default settings. SSU sequences from each MAG were then re-parsed and annotated by SINA v.1.2.11 [ 102 ]. Scaffolds containing 16S rRNA gene sequences inconsistent with GTDB taxonomic classifications were deemed contaminants and were removed. Selected MAGs were then further refined and manually inspected by VizBin v.1.0.0 [ 103 ]. Final MAGs had an estimated ≥ 50% genome completion and ≤ 10% contamination, with completeness and contamination rounded to the nearest whole number.
Iterative Nanoarchaeota MAG curation
As a case study, two MAGs assigned to the Nanoarchaeota (4571-419_metabat1_scaf2bin.008, M10_maxbin2_scaf2bin.065) were iteratively curated, demonstrating that the original MAGs generated by DAS Tool contained large quantities of contaminant contigs that were not recognized by CheckM, given the low abundance of marker genes. Each MAG was visualized using the Anvi’o v.7.1 interactive interface [ 104 ], where contigs were divided into subsets based on clustering patterns in Anvi’o. Contigs in each cluster were assigned a putative taxonomy using the Contig Annotation Tool (CAT) [ 105 ]. Clusters containing most of the contigs assigned to the Nanoarchaeota were repeatedly sub-sampled and screened using the CAT pipeline until no meaningful correspondence between clustering patterns and assigned taxonomy could be identified (Fig. S 5 ). Contigs in the final clusters were then removed if CAT definitively assigned them to a taxonomic group outside the Nanoarchaeota, while contigs assigned to the Nanoarchaeota and unclassified higher ranks were retained. A third Nanoarchaeota MAG (4281-140_maxbin2_scaf2bin.078) was also identified, but attempted curation using the above workflow revealed the presence of extensive contamination, with only a very small subset of scaffolds confidently assigned to the Nanoarchaeota. CAT analysis of a putative Nanoarchaeota MAG (JGI Bin ID 3300028417_39) separately assembled from the same read set by the JGI as part of the Genomes from Earth’s Microbiomes project [ 106 ] also showed very few contigs assigned to the DPANN superphylum and extensive bacterial contamination, suggesting that this particular read set may represent a challenge for commonly utilized binning algorithms. Given the extensive contamination and difficulty identifying a valid Nanoarchaeota MAG of significant size, the 4281-140_maxbin2_scaf2bin.078 was excluded from the MAG dataset submitted to Genbank, so as to avoid contaminating the public database with erroneous information. However, the MAG was included in functional and relative abundance calculations.
MAG characterization and annotation
Open reading frames (ORFs) were predicted by Prodigal v.2.6.3 [ 107 ] with the parameter “-p meta”. ORFs were then annotated by KOfam [ 108 ] and custom HMM profiles within METABOLIC v.4.0 [ 109 ] and eggNOG-emapper v.2.1.2 [ 110 ] with default settings. Transfer RNAs were predicted using tRNAscan-SE 2.0 using the general tRNA model [ 111 ]. Genomic properties, including genome coverage, genome and 16S rRNA taxonomy, tRNAs, genome completeness, and scaffold parameters, were parsed from results that were calculated by CheckM, tRNAscan-SE 2.0, and METABOLIC. Relative genome coverages were normalized by setting each metagenomic dataset size as 100 M paired-end reads.
Prior to detailed metabolic analysis, open reading frames from the Gracilibacteria orders BD1-5 and Absconditabacterales, which are known to use genetic code 25 (e.g., [ 47 , 48 , 112 , 113 ]), were re-called using Prodigal v.2.6.3 as implemented in Prokka v.1.14.6 [ 114 ]. An additional MAG from the Gracilibacteria order GCA-2401425 (4559-240_metabat1_scaf2bin.085) was also processed using genetic code 25. Currently, the only other genome in GTDB order GCA-2401425 (Genbank accession NVTB00000000.1) [ 115 ] is publicly available in Genbank with ORFs generated using genetic code 11. However, comparative analysis of our GCA-2401425 MAG showed that ORFs called with genetic code 11 were truncated, with an average length of approximately 85 amino acids, while those called with genetic code 25 averaged 277 amino acids in length. ORFs from two additional MAGs from the Paceibacteria (A3_metabat2_scaf2bin.333 and S145_metabat2_scaf2bin.004) were also re-generated in Prokka using genetic code 11. Open reading frames were then annotated in GhostKoala [ 116 ].
Phylogenomic inference
For archaeal phylogenomic tree construction, a concatenated multiple sequence alignment (MSA) was generated in GTDB-Tk using 122 archaeal marker genes (2991 sequences, 5124 columns) [ 17 ]. IQ-TREE v.1.6.9 [ 117 ] was used to reconstruct the tree with the settings “-m MFP -bb 1000 -redo -mset WAG,LG,JTT,Dayhoff -mrate E,I,G,I + G -mfreq FU -wbtl” (Data S 1 ). The bacterial phylogenomic tree was constructed in a similar manner, using a concatenated MSA of 120 bacterial GTDB marker genes [ 17 ]. For each GTDB bacterial phylum, no more than 15 reference genomes from the GTDB r202 database were used (4248 sequences, 5037 columns; Data S 2 ). Additionally, a second bacterial phylogenomic tree was inferred from the same MSA using FastTree v.2.1.8 (WAG, + gamma, SH support; Data S 3 ) [ 118 ]. Additional MSAs solely using MAGs from this study were generated for the Archaea (122 marker genes) and Bacteria (120 marker genes) using the GTDB-Tk identify and align commands [ 17 ]. FastTree v.2.1.10 (parameter: –gamma) was used to infer the phylogenomic trees, as implemented in GTDB-Tk (Data S 4 , S 5 ; formatted trees available online at https://itol.embl.de/shared/alrlab ).
A tree was constructed in GTDB-Tk (parameter: –gamma) using MAGs assigned to the Patescibacteria, along with recently described Cand . Vampirococcus lugosii [ 47 ] and Cand . Absconditicoccus praedator [ 48 ], and the GTDB r202 bacterial tree-building dataset. A phylogenomic tree of the Chloroflexota was also generated by extracting a concatenated MSA of Chlorofexota MAGs from the entire bacterial MSA. IQ-TREE v.2.1.4 [ 119 ] was used to reconstruct the tree with the settings “-m TESTMERGE -bb 1000 -bnni”. An outgroup genome (GCA_007123655.1) was added to reroot the phylogenomic tree. Final trees were visualized using Interactive Tree of Life (iTOL) v.6 [ 120 ].
Taxonomic assignment
Initial taxonomy was assigned to each MAG using the GTDB-Tk classify pipeline. In rare instances where there were discrepancies between the class-level (Archaea) or phylum-level taxonomy (Bacteria) assigned by GTDB-Tk and phylogenetic tree topology, we deferred to tree topology. In the Bacteria, topological taxonomic assignments were only used if confirmed by both trees. MAGs that were not assigned to a known genus by GTDB-Tk were compared to their closest relatives in this study using average amino acid identity (AAI) matrices generated in CompareM v.0.1.2 ( https://github.com/dparks1134/CompareM ). MAGs were assigned to novel genera using cutoffs provided by Konstantinidis et al. [ 18 ], and MAGs assigned the taxonomic status “unclassified” were automatically assigned to a novel genus.
Trophic and energy metabolism analysis
Functional genes were first characterized by METABOLIC [ 109 ]. Additional peptide utilization genes were characterized using the MEROPS database release 12.3 [ 121 ], and additional polysaccharide utilization genes were identified using dbCAN2 (2020–04-08) and the CAZy (2021–05-31) database [ 122 , 123 ]. Cellular localization of peptidases/inhibitors, gene calls identified by the CAZy database, and predicted extracellular nucleases were verified using PSORTb v.3.0 [ 124 ]. Functional annotations for protein, polysaccharide, nucleic acid, and lipid utilization were derived in part from previous publications [ 125 , 126 ]. Iron cycling genes and hydrogenase genes were characterized based on HMMs directly obtained or indirectly parsed from FeGenie [ 127 ] and HydDB [ 75 ].
For each of these trophic and energy metabolisms, the number of functional gene calls in each genome was calculated using two different scenarios: (1) the presence of any marker gene in the complex/pathway was treated as the presence of the whole function (indicated as C), and the highest number of gene calls for an individual gene in the complex was taken to be the number of pathway “hits” in the MAG. (2) Stand-alone genes that were not part of a large complex or functional pathway (indicated as A) were treated as individual accumulative gene calls for their particular function. In specific cases, marker genes were manually verified using phylogenetic trees and by inspecting operon arrangements (see below). To calculate functional abundance, all genomes were included in the analysis. Functional abundance was then calculated by multiplying normalized genome coverage (100 M reads/sample) by the number of functional gene calls for each sample. For visualization, functional abundance was then log-transformed and used to generate heatmaps with the R package pheatmap v.1.0.12 (settings: clustering_method = ward.D2). Combined functional heatmaps were also generated by summing values within larger functional groups.
To avoid potential mis-annotation by the automated methods described above, phylogenetic trees were constructed to validate predicted protein sequences for dissimilatory sulfite reductase (Dsr; Fig. S 9 ), methyl-coenzyme M reductase subunit alpha (McrA; Fig. S 10 ), and sulfur dioxygenase (Sdo; Fig. S 11 ). Based on current understanding, two metabolic directions are possible for the Dsr protein: reductive Dsr, which catalyzes the reduction of sulfite to sulfide, and oxidative (or reverse) Dsr, which converts elemental sulfur oxidation to sulfite [ 128 ]. Paired DsrAB proteins were first identified in all MAGs using in-house Perl scripts. In cases where Dsr subunits were duplicated, one set of paired DsrAB proteins was manually selected. A concatenated protein alignment was then generated for DsrAB proteins from the MAGs and reference sequences using MAFFT v.7.310 [ 129 ], and the alignment was trimmed using trimAl v.1.4.rev15 [ 130 ] with the parameter “-gt 0.25”. A phylogenetic tree was then constructed in IQ-TREE with settings “-m MFP -bb 1000 -redo -mset WAG,LG,JTT,Dayhoff -mrate E,I,G,I + G -mfreq FU -wbtl” (Fig. S 9 ). Reductive and oxidative DsrAB proteins were identified based on placement in the phylogenetic tree.
Predicted proteins for McrA were first identified using the TIGR03256 HMM. Presumed false gene calls were then manually removed, including those identified in bacterial MAGs and non-methanogenic/anaerobic methanotrophic archaeal MAGs with high sequence coverage. An alignment was constructed in MAFFT v.7.310 [ 129 ] using the remaining McrA protein sequences, together with reference genes recovered from methanogens, anaerobic methanotrophs, and short-chain alkane oxidizing Archaea from the Bathyarchaeia, Helarchaeales, Syntrophoarchaeum and Polytropus [ 11 , 12 , 131 , 132 ]. Alignment trimming and phylogenetic tree inference were performed as described above.
Sulfur dioxygenase (Sdo) proteins were predicted using the “sulfur_dioxygenase_sdo” HMM [ 109 ]. Alignment, trimming, and construction of the phylogeny were performed as described above. Positive Sdo calls were identified using two conserved amino acid residues (Asp196 and Asn244 of hETHE1, NCBI accession NP_055112) that are specific to Sdo in comparison with other metallo-β-lactamase superfamily members [ 133 ].
Statistical analysis
The relative abundance of MAGs in this study was calculated for each sample using normalized read coverage (set to 100 M reads) expressed as a percentage. Bray–Curtis similarity matrices were then generated from relative abundance data at various taxonomic ranks, and nonmetric multidimensional scaling (NMDS) plots were generated from the matrices using PRIMER v.6.1.13 [ 134 ].
Availability of data and materials
Metagenome reads are publicly available in the Sequence Read Archive (Table S 1 ), and MAGs generated in this study are available in NCBI Genbank (BioProject PRJNA821212, Table S 2 ).
Nakagawa S, Takai K, Inagaki F, Chiba H, Ishibashi JI, Kataoka S, et al. Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation. FEMS Microbiol Ecol. 2005;54:141–55.
Article CAS Google Scholar
Nunoura T, Takai K. Comparison of microbial communities associated with phase-separation- induced hydrothermal fluids at the Yonaguni Knoll IV hydrothermal field, the Southern Okinawa Trough. FEMS Microbiol Ecol. 2009;67:351–70.
Flores GE, Campbell JH, Kirshtein JD, Meneghin J, Podar M, Steinberg JI, et al. Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ Microbiol. 2011;13:2158–71.
Flores GE, Shakya M, Meneghin J, Yang ZK, Seewald JS, Geoff Wheat C, et al. Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology. 2012;10:333–46.
Dahle H, Økland I, Thorseth IH, Pederesen RB, Steen IH. Energy landscapes shape microbial communities in hydrothermal systems on the Arctic Mid-Ocean Ridge. ISME J. 2015;9:1593–606.
Dahle H, Le Moine BS, Baumberger T, Stokke R, Pedersen RB, Thorseth IH, et al. Energy landscapes in hydrothermal chimneys shape distributions of primary producers. Front Microbiol. 2018;9:1570.
Article Google Scholar
Fortunato CS, Larson B, Butterfield DA, Huber JA. Spatially distinct, temporally stable microbial populations mediate biogeochemical cycling at and below the seafloor in hydrothermal vent fluids. Environ Microbiol. 2018;20:769–84.
Reysenbach A-L, St. John E, Meneghin J, Flores GE, Podar M, Dombrowski N, et al. Complex subsurface hydrothermal fluid mixing at a submarine arc volcano supports distinct and highly diverse microbial communities. Proc Natl Acad Sci U S A. 2020;117:32627–38.
Reveillaud J, Reddington E, McDermott J, Algar C, Meyer JL, Sylva S, et al. Subseafloor microbial communities in hydrogen-rich vent fluids from hydrothermal systems along the Mid-Cayman Rise. Environ Microbiol. 2016;18:1970–87.
Anderson RE, Reveillaud J, Reddington E, Delmont TO, Eren AM, McDermott JM, et al. Genomic variation in microbial populations inhabiting the marine subseafloor at deep-sea hydrothermal vents. Nat Commun. 2017;8:1114.
Dombrowski N, Seitz KW, Teske AP, Baker BJ. Genomic insights into potential interdependencies in microbial hydrocarbon and nutrient cycling in hydrothermal sediments. Microbiome. 2017;5:106.
Dombrowski N, Teske AP, Baker BJ. Expansive microbial metabolic versatility and biodiversity in dynamic Guaymas Basin hydrothermal sediments. Nat Commun. 2018;9:4999.
Ramírez GA, McKay LJ, Fields MW, Buckley A, Mortera C, Hensen C, et al. The Guaymas Basin subseafloor sedimentary archaeome reflects complex environmental histories. iScience. 2020;23:101459.
Xie W, Wang F, Guo L, Chen Z, Sievert SM, Meng J, et al. Comparative metagenomics of microbial communities inhabiting deep-sea hydrothermal vent chimneys with contrasting chemistries. ISME J. 2011;5:414–26.
Hou J, Sievert SM, Wang Y, Seewald JS, Natarajan VP, Wang F, et al. Microbial succession during the transition from active to inactive stages of deep-sea hydrothermal vent sulfide chimneys. Microbiome. 2020;8:102.
Meier DV, Pjevac P, Bach W, Hourdez S, Girguis PR, Vidoudez C, et al. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017;11:1545–58.
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics. 2020;36:1925–7.
CAS Google Scholar
Konstantinidis KT, Rosselló-Móra R, Amann R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017;11:2399–406.
Von Damm KL, Edmond JM, Measures CI, Grant B. Chemistry of submarine hydrothermal solutions at Guaymas Basin, Gulf of California. Geochim Cosmochim Acta. 1985;49:2221–37.
Teske A, de Beer D, McKay LJ, Tivey MK, Biddle JF, Hoer D, et al. The Guaymas Basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: complex seafloor expressions of subsurface hydrothermal circulation. Front Microbiol. 2016;7:75.
Burggraf S, Larsen N, Woese CR, Stetter KO. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum . Proc Natl Acad Sci U S A. 1993;90:2547–50.
Nomura N, Morinaga Y, Kogishi T, Kim E-J, Sako Y, Uchida A. Heterogeneous yet similar introns reside in identical positions of the rRNA genes in natural isolates of the archaeon Aeropyrum pernix . Gene. 2002;295:43–50.
Jay ZJ, Inskeep WP. The distribution, diversity, and importance of 16S rRNA gene introns in the order Thermoproteales. Biol Direct. 2015;10:35.
St. John E, Liu Y, Podar M, Stott MB, Meneghin J, Chen Z, et al. A new symbiotic nanoarchaeote ( Candidatus Nanoclepta minutus) and its host ( Zestosphaera tikiterensis gen. nov., sp. Nov.) from a New Zealand hot spring. Syst Appl Microbiol. 2019;42:94–106.
Reysenbach A-L, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, et al. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–7.
Kozubal MA, Romine M, Jennings RD, Jay ZJ, Tringe SG, Rusch DB, et al. Geoarchaeota: a new candidate phylum in the Archaea from high-temperature acidic iron mats in Yellowstone National Park. ISME J. 2013;7:622–34.
McKay L, Klokman VW, Mendlovitz HP, Larowe DE, Hoer DR, Albert D, et al. Thermal and geochemical influences on microbial biogeography in the hydrothermal sediments of Guaymas Basin, Gulf of California. Environ Microbiol Rep. 2016;8:150–61.
Zhou Z, Liu Y, Xu W, Pan J, Luo Z-H, Li M. Genome- and community-level interaction insights into carbon utilization and element cycling functions of Hydrothermarchaeota in hydrothermal sediment. mSystems. 2020;5:e00795-19.
Buessecker S, Palmer M, Lai D, Dimapilis J, Mayali X, Mosier D, et al. An essential role for tungsten in the ecology and evolution of a previously uncultivated lineage of anaerobic, thermophilic Archaea. Nat Commun. 2022;13:3773.
Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048.
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004.
Luef B, Frischkorn KR, Wrighton KC, Holman H-YN, Birarda G, Thomas BC, et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 2015;6:6372.
Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, et al. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio. 2013;4:e00708-e713.
Castelle CJ, Brown CT, Anantharaman K, Probst AJ, Huang RH, Banfield JF. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol. 2018;16:629–45.
Tian R, Ning D, He Z, Zhang P, Spencer SJ, Gao S, et al. Small and mighty: adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome. 2020;8:51.
Lemos LN, Manoharan L, Mendes LW, Venturini AM, Pylro VS, Tsai SM. Metagenome assembled-genomes reveal similar functional profiles of CPR/Patescibacteria phyla in soils. Environ Microbiol Rep. 2020;12:651–5.
Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, et al. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun. 2016;7:13219.
McLean JS, Bor B, Kerns KA, Liu Q, To TT, Solden L, et al. Acquisition and adaptation of ultra-small parasitic reduced genome Bacteria to mammalian hosts. Cell Rep. 2020;32:107939.
Cross KL, Campbell JH, Balachandran M, Campbell AG, Cooper SJ, Griffen A, et al. Targeted isolation and cultivation of uncultivated bacteria by reverse genomics. Nat Biotechnol. 2019;37:1314–21.
He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu S-Y, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A. 2015;112:244–9.
Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–7.
Anantharaman K, Breier JA, Dick GJ. Metagenomic resolution of microbial functions in deep-sea hydrothermal plumes across the Eastern Lau Spreading Center. ISME J. 2016;10:225–39.
Beam JP, Becraft ED, Brown JM, Schulz F, Jarett JK, Bezuidt O, et al. Ancestral absence of electron transport chains in Patescibacteria and DPANN. Front Microbiol. 2020;11:1848.
Kato S, Nakawake M, Kita J, Yamanaka T, Utsumi M, Okamura K, et al. Characteristics of microbial communities in crustal fluids in a deep-sea hydrothermal field of the Suiyo Seamount. Front Microbiol. 2013;4:85.
Oulas A, Polymenakou PN, Seshadri R, Tripp HJ, Mandalakis M, Paez-Espino AD, et al. Metagenomic investigation of the geologically unique Hellenic Volcanic Arc reveals a distinctive ecosystem with unexpected physiology. Environ Microbiol. 2016;18:1122–36.
Sieber CMK, Paul BG, Castelle CJ, Hu P, Tringe SG, Valentine DL, et al. Unusual metabolism and hypervariation in the genome of a gracilibacterium (BD1-5) from an oil-degrading community. MBio. 2019;10:e02128-e2219.
Moreira D, Zivanovic Y, López-Archilla AI, Iniesto M, López-García P. Reductive evolution and unique predatory mode in the CPR bacterium Vampirococcus lugosii . Nat Commun. 2021;12:2454.
Yakimov MM, Merkel AY, Gaisin VA, Pilhofer M, Messina E, Hallsworth JE, et al. Cultivation of a vampire: ‘ Candidatus Absconditicoccus praedator’. Environ Microbiol. 2022;24:30–49.
Coutinho FH, von Meijenfeldt FAB, Walter JM, Haro-Moreno JM, Lopéz-Pérez M, van Verk MC, et al. Ecogenomics and metabolic potential of the South Atlantic Ocean microbiome. Sci Total Environ. 2021;765:142758.
Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, et al. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome. 2013;1:22.
Fincker M, Huber JA, Orphan VJ, Rappé MS, Teske A, Spormann AM. Metabolic strategies of marine subseafloor Chloroflexi inferred from genome reconstructions. Environ Microbiol. 2020;22:3188–204.
Fullerton H, Moyer CL. Comparative single-cell genomics of Chloroflexi from the Okinawa Trough deep-subsurface biosphere. Appl Environ Microbiol. 2016;82:3000–8.
Nuppunen-Puputti M, Kietäväinen R, Raulio M, Soro A, Purkamo L, Kukkonen I, et al. Epilithic microbial community functionality in deep oligotrophic continental bedrock. Front Microbiol. 2022;13:826048.
West-Roberts JA, Matheus-Carnevali PB, Schoelmerich MC, Al-Shayeb B, Thomas AD, Sharrar A, et al. The Chloroflexi supergroup is metabolically diverse and representatives have novel genes for non-photosynthesis based CO 2 fixation. BioRxiv. https://doi.org/10.1101/2021.08.23.457424 .
Liu R, Wei X, Song W, Wang L, Cao J, Wu J, et al. Novel Chloroflexi genomes from the deepest ocean reveal metabolic strategies for the adaptation to deep-sea habitats. Microbiome. 2022;10:75.
McGonigle JM, Lang SQ, Brazelton WJ. Genomic evidence for formate metabolism by Chloroflexi as the key to unlocking deep carbon in Lost City microbial ecosystems. Appl Environ Microbiol. 2020;86:e02583-e2619.
Islam ZF, Cordero PRF, Feng J, Chen Y-J, Bay SK, Jirapanjawat T, et al. Two Chloroflexi classes independently evolved the ability to persist on atmospheric hydrogen and carbon monoxide. ISME J. 2019;13:1801–13.
Altshuler I, Raymond-Bouchard I, Magnuson E, Tremblay J, Greer CW, Whyte LG. Unique high Arctic methane metabolizing community revealed through in situ 13 CH 4- DNA-SIP enrichment in concert with genome binning. Sci Rep. 2022;12:1160.
Madigan MT, Brock TD. Photosynthetic sulfide oxidation by Chloroflexus aurantiacus , a filamentous, photosynthetic, gliding bacterium. J Bacteriol. 1975;122:782–4.
Kawai S, Martinez JN, Lichtenberg M, Trampe E, Kühl M, Tank M, et al. In-situ metatranscriptomic analyses reveal the metabolic flexibility of the thermophilic anoxygenic photosynthetic bacterium Chloroflexus aggregans in a hot spring Cyanobacteria-dominated microbial mat. Microorganisms. 2021;9:652.
Spieck E, Spohn M, Wendt K, Bock E, Shively J, Frank J, et al. Extremophilic nitrite-oxidizing Chloroflexi from Yellowstone hot springs. ISME J. 2020;14:364–79.
Baker BJ, Lesniewski RA, Dick GJ. Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J. 2012;6:2269–79.
Engelen B, Nguyen T, Heyerhoff B, Kalenborn S, Sydow K, Tabai H, et al. Microbial communities of hydrothermal Guaymas Basin surficial sediment profiled at 2 millimeter-scale resolution. Front Microbiol. 2021;12:710881.
Beman JM, Popp BN, Francis CA. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2008;2:429–41.
Speth DR, Yu FB, Connon SA, Lim S, Magyar JS, Peña-Salinas ME, et al. Microbial communities of Auka hydrothermal sediments shed light on vent biogeography and the evolutionary history of thermophily. ISME J. 2022;16:1750–64.
Mehta MP, Baross JA. Nitrogen fixation at 92°C by a hydrothermal vent archaeon. Science. 2006;314:1783–6.
Dick GJ. The microbiomes of deep-sea hydrothermal vents: distributed globally, shaped locally. Nat Rev Microbiol. 2019;17:271–83.
Frank KL, Rogers DR, Olins HC, Vidoudez C, Girguis PR. Characterizing the distribution and rates of microbial sulfate reduction at Middle Valley hydrothermal vents. ISME J. 2013;7:1391–401.
Zhou Z, Tran PQ, Adams AM, Kieft K, Breier JA, Sinha RK, et al. The sulfur cycle connects microbiomes and biogeochemistry in deep-sea hydrothermal plumes. BioRxiv. https://doi.org/10.1101/2022.06.02.494589 .
Von Damm KL, Parker CM, Zierenberg RA, Lilley MD, Olson EJ, Clague DA, et al. The Escanaba Trough, Gorda Ridge hydrothermal system: temporal stability and subseafloor complexity. Geochim Cosmochim Acta. 2005;69:4971–84.
Dhillon A, Lever M, Lloyd KG, Albert DB, Sogin ML, Teske A. Methanogen diversity evidenced by molecular characterization of methyl coenzyme M reductase A ( mcrA ) genes in hydrothermal sediments of the Guaymas Basin. Appl Environ Microbiol. 2005;71:4592–601.
Adam N, Perner M. Microbially mediated hydrogen cycling in deep-sea hydrothermal vents. Front Microbiol. 2018;9:2873.
Lever MA, Teske AP. Diversity of methane-cycling Archaea in hydrothermal sediment investigated by general and group-specific PCR primers. Appl Environ Microbiol. 2015;81:1426–41.
Teske A, Wegener G, Chanton JP, White D, MacGregor B, Hoer D, et al. Microbial communities under distinct thermal and geochemical regimes in axial and off-axis sediments of Guaymas Basin. Front Microbiol. 2021;12:633649.
Søndergaard D, Pedersen CNS, Greening C. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:34212.
Kodama Y, Watanabe K. Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int J Syst Evol Microbiol. 2004;54:2297–300.
Takai K, Inagaki F, Nakagawa S, Hirayama H, Nunoura T, Sako Y, et al. Isolation and phylogenetic diversity of members of previously uncultivated ε-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol Lett. 2003;218:167–74.
Takai K, Suzuki M, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F, et al. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas . Int J Syst Evol Microbiol. 2006;56:1725–33.
Caldwell SL, Liu Y, Ferrera I, Beveridge T, Reysenbach A-L. Thermocrinis minervae sp. nov., a hydrogen- and sulfur-oxidizing, thermophilic member of the Aquificales from a Costa Rican terrestrial hot spring. Int J Syst Evol Microbiol. 2010;60:338–43.
Götz D, Banta A, Beveridge TJ, Rushdi AI, Simoneit BRT, Reysenbach A-L. Persephonella marina gen. nov., sp. nov. and Persephonella guaymasensis sp. nov., two novel, thermophilic, hydrogen-oxidizing microaerophiles from deep-sea hydrothermal vents. Int J Syst Evol Microbiol. 2002;52:1349–59.
Google Scholar
Topçuoglu BD, Stewart LC, Morrison HG, Butterfield DA, Huber JA, Holden JF. Hydrogen limitation and syntrophic growth among natural assemblages of thermophilic methanogens at deep-sea hydrothermal vents. Front Microbiol. 2016;7:1240.
Webster NS. Cooperation, communication, and co-evolution: grand challenges in microbial symbiosis research. Front Microbiol. 2014;5:164.
Petersen JM, Zielinski FU, Pape T, Seifert R, Moraru C, Amann R, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–80.
Galambos D, Anderson RE, Reveillaud J, Huber JA. Genome-resolved metagenomics and metatranscriptomics reveal niche differentiation in functionally redundant microbial communities at deep-sea hydrothermal vents. Environ Microbiol. 2019;21:4395–410.
Louca S, Parfrey LW, Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353:1272–7.
Yamamoto M, Takai K. Sulfur metabolisms in Epsilon- and Gamma-proteobacteria in deep-sea hydrothermal fields. Front Microbiol. 2011;2:192.
Giovannelli D, Chung M, Staley J, Starovoytov V, Le Bris N, Vetriani C. Sulfurovum riftiae sp. nov., a mesophilic, thiosulfate-oxidizing, nitrate-reducing chemolithoautotrophic epsilonproteobacterium isolated from the tube of the deep-sea hydrothermal vent polychaete Riftia pachyptila . Int J Syst Evol Microbiol. 2016;66:2697–701.
Mori K, Yamaguchi K, Hanada S. Sulfurovum denitrificans sp. nov., an obligately chemolithoautotrophic sulfur-oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int J Syst Evol Microbiol. 2018;68:2183–7.
Nakagawa S, Takai K, Inagaki F, Horikoshi K, Sako Y. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the ε-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int J Syst Evol Microbiol. 2005;55:925–33.
Assié A, Leisch N, Meier DV, Gruber-Vodicka H, Tegetmeyer HE, Meyerdierks A, et al. Horizontal acquisition of a patchwork Calvin cycle by symbiotic and free-living Campylobacterota (formerly Epsilonproteobacteria). ISME J. 2020;14:104–22.
Berg IA. Ecological aspects of the distribution of different autotrophic CO 2 fixation pathways. Appl Environ Microbiol. 2011;77:1925–36.
Markert S, Arndt C, Felbeck H, Becher D, Sievert SM, Hügler M, et al. Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila . Science. 2007;315:247–50.
Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K, et al. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front Microbiol. 2017;8:682.
Macalady JL, Dattagupta S, Schaperdoth I, Jones DS, Druschel GK, Eastman D. Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J. 2008;2:590–601.
Patwardhan S, Foustoukos DI, Giovannelli D, Yücel M, Vetriani C. Ecological succession of sulfur-oxidizing Epsilon- and Gammaproteobacteria during colonization of a shallow-water gas vent. Front Microbiol. 2018;9:2970.
Flores GE, Wagner ID, Liu Y, Reysenbach A-L. Distribution, abundance, and diversity patterns of the thermoacidophilic “deep-sea hydrothermal vent Euryarchaeota 2.” Front Microbiol. 2012;3:47.
Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34.
Uritskiy GV, DiRuggiero J, Taylor J. MetaWRAP – a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158.
Sieber CMK, Probst AJ, Sharrar A, Thomas BC, Hess M, Tringe SG, et al. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat Microbiol. 2018;3:836–43.
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55.
Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42.
Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9.
Laczny CC, Sternal T, Plugaru V, Gawron P, Atashpendar A, Margossian HH, et al. VizBin - an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome. 2015;3:1.
Eren AM, Kiefl E, Shaiber A, Veseli I, Miller SE, Schechter MS, et al. Community-led, integrated, reproducible multi-omics with Anvi’o. Nat Microbiol. 2021;6:3–6.
von Meijenfeldt FAB, Arkhipova K, Cambuy DD, Coutinho FH, Dutilh BE. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT. Genome Biol. 2019;20:217.
Nayfach S, Roux S, Seshadri R, Udwary D, Varghese N, Schulz F, et al. A genomic catalog of Earth’s microbiomes. Nat Biotechnol. 2021;39:499–509.
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119.
Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, et al. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2.
Zhou Z, Tran PQ, Breister AM, Liu Y, Kieft K, Cowley ES, et al. METABOLIC: high-throughput profiling of microbial genomes for functional traits, metabolism, biogeochemistry, and community-scale functional networks. Microbiome. 2022;10:33.
Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47:D309-14.
Chan PP, Lowe TM. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 2019;1962:1–14.
Campbell JH, O’Donoghue P, Campbell AG, Schwientek P, Sczyrba A, Woyke T, et al. UGA is an additional glycine codon in uncultured SR1 Bacteria from the human microbiota. Proc Natl Acad Sci U S A. 2013;110:5540–5.
Hanke A, Hamann E, Sharma R, Geelhoed JS, Hargesheimer T, Kraft B, et al. Recoding of the stop codon UGA to glycine by a BD1-5/SN-2 bacterium and niche partitioning between Alpha- and Gammaproteobacteria in a tidal sediment microbial community naturally selected in a laboratory chemostat. Front Microbiol. 2014;5:231.
Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9.
Tully BJ, Wheat CG, Glazer BT, Huber JA. A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 2018;12:1–16.
Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–31.
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74.
Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490.
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4.
Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:W293–6.
Rawlings ND, Barrett AJ, Thomas PD, Huang X, Bateman A, Finn RD. The MEROPS database of proteolytic enzymes, their substrates and inhibitors in 2017 and a comparison with peptidases in the PANTHER database. Nucleic Acids Res. 2018;46:D624–32.
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, et al. DbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95-101.
Drula E, Garron M-L, Dogan S, Lombard V, Henrissat B, Terrapon N. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 2022;50:D571–7.
Yu NY, Wagner JR, Laird MR, Melli G, Rey S, Lo R, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–15.
Shaffer M, Borton MA, McGivern BB, Zayed AA, La Rosa SL, Solden LM, et al. DRAM for distilling microbial metabolism to automate the curation of microbiome function. Nucleic Acids Res. 2020;48:8883–900.
Pérez Castro S, Borton MA, Regan K, Hrabe de Angelis I, Wrighton KC, Teske AP, et al. Degradation of biological macromolecules supports uncultured microbial populations in Guaymas Basin hydrothermal sediments. ISME J. 2021;15:3480–97.
Garber AI, Nealson KH, Okamoto A, McAllister SM, Chan CS, Barco RA, et al. FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol. 2020;11:37.
Anantharaman K, Hausmann B, Jungbluth SP, Kantor RS, Lavy A, Warren LA, et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018;12:1715–28.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3.
Seitz KW, Dombrowski N, Eme L, Spang A, Lombard J, Sieber JR, et al. Asgard Archaea capable of anaerobic hydrocarbon cycling. Nat Commun. 2019;10:1822.
Boyd JA, Jungbluth SP, Leu AO, Evans PN, Woodcroft BJ, Chadwick GL, et al. Divergent methyl-coenzyme M reductase genes in a deep-subseafloor Archaeoglobi. ISME J. 2019;13:1269–79.
Liu H, Xin Y, Xun L. Distribution, diversity, and activities of sulfur dioxygenases in heterotrophic bacteria. Appl Environ Microbiol. 2014;80:1799–806.
Clarke KR, Gorley RN. Primer V6: user manual - tutorial. Plymouth: Plymouth Marine Laboratory; 2006.
Download references
Acknowledgements
We thank the crew of the R/V Roger Revelle , R/V Atlantis , R/V Thomas G. Thompson , HOV Alvin , and the ROV Jason for assistance in collecting the samples. Many thanks to the many students who over the years helped extract the DNA, to Nicole Wagner and Jennifer Meneghin for initial bioinformatic analysis assistance, and to MK Tivey for thoughtful comments on the manuscript.
This work was funded by the US-National Science Foundation grants OCE-0728391, OCE-0937404, OCE-1558795 to A-L.R, and OCE-2049478 and DBI-2047598 to K.A. We thank the Department of Energy Joint Genome Institute (Community Science Program award 339, lead Peter Girguis) for sequencing several of the samples.
Author information
Zhichao Zhou and Emily St. John contributed equally to this work.
Authors and Affiliations
Department of Bacteriology, University of Wisconsin-Madison, Madison, WI, 53706, USA
Zhichao Zhou & Karthik Anantharaman
Center for Life in Extreme Environments, Biology Department, Portland State University, Portland, OR, 97201, USA
Emily St. John & Anna-Louise Reysenbach
You can also search for this author in PubMed Google Scholar
Contributions
A-L.R conceived of the study, collected and processed the samples, and wrote the manuscript; Z.Z. and E.S.J. did the bioinformatic processing and data analysis and generated the figures and tables; K.A. assisted in project conception and data analysis; and all authors read, reviewed, edited, and approved the final manuscript.
Corresponding authors
Correspondence to Karthik Anantharaman or Anna-Louise Reysenbach .
Ethics declarations
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: fig. s1..
Geographic distribution of deep-sea hydrothermal vent sampling locations. The number of samples collected in each region is shown with n values. Fig. S2. Deep-sea hydrothermal vent photographs from ELSC, EPR, MAR and Guaymas Basin. Fig. S3. Comparison between the number of medium- to high-quality MAGs recovered in each metagenomic assembly and the number of reads that passed quality control measures. Metagenomic assemblies are ordered by (A) increasing MAG count and (B) increasing read count. Fig. S4. NMDS plot showing the taxonomic diversity of Brothers volcano MAGs, based on normalized relative abundance. Clustering patterns show a high degree of similarity to NMDS plot clustering previously reported in Reysenbach et al., 2020. Fig. S5. Anvi’o plot showing the cluster of scaffolds (blue) predominantly corresponding to the Nanoarchaeota in M10_maxbin2_scaf2bin.065. Analysis with CAT revealed three additional contaminating scaffolds which were removed, bringing the final scaffold count to 149, with an estimated 47% completion by CheckM. Scaffold clusters that were removed (pink; 972 scaffolds) were largely assigned to taxonomic groups outside the Nanoarchaeota by CAT and had a low number of marker genes, as estimated by CheckM (6.99% completion, 0.29% contamination). Fig. S6. Predicted cell metabolism diagrams for the putative new phyla (A) JALSQH01 (3 MAGs) and (B) JALWCF01 (13 MAGs). Functions (F) and modules (M) were identified using METABOLIC (Table S5). Solid lines indicate the presence of a module or function, while dashed lines and a “p” in parentheses indicate that a module or function was only present sporadically (<50% of MAGs). Modules and functions not identified in any MAGs are shown with dashed lines and gray labels. Fig. S7. Normalized relative abundance of GTDB classes, expressed as a percentage. Classes depicted comprise ≥16% of the relative MAG abundance in at least one assembly. Fig. S8. Maximum-likelihood GTDB-Tk concatenated protein tree showing members of the Patescibacteria, used to generate Fig. 5A. Lineages outside the Patescibacteria are shown as a collapsed triangle, and MAGs from this study are indicated in bold type. Filled circles represent SH-like branch support (0.8-1.0), and the scale bar shows 0.5 substitutions per amino acid. Fig. S9. Concatenated dissimilatory sulfite reductase (DsrAB) protein phylogenetic tree. Only the nodes with ultrafast bootstrap (UFBoot) support values over 90% were labeled with black dots. This tree included both reductive DsrAB (for reductive dissimilatory sulfite reduction to sulfide) and oxidative DsrAB (for dissimilatory sulfur oxidation to sulfite). For collapsed clades in the oxidative DsrAB clade (labeled in blue), the DsrAB call numbers and DsrAB-containing MAG numbers were labeled in square brackets. The total number for both reductive DsrAB calls and reductive DsrAB-containing MAG numbers and oxidative DsrAB calls and oxidative DsrAB-containing MAG numbers were labeled accordingly on the side of the tree. Note that one genome can have multiple paired DsrAB calls. Fig. S10. Phylogenetic protein tree of methyl coenzyme M reductase subunit alpha (McrA). Ultrafast bootstrap support values (>90%) are shown with filled circles. Clades comprised of predicted butane oxidation (Butane clade), X-alkane oxidation (X-alkane clade) and anaerobic methanotrophy-associated (ANME-1 and -2) McrA amino acid sequences are highlighted, and the three predicted McrA sequences from the Archaeoglobi are shown in red. Fig. S11. Sdo (sulfur dioxygenase) phylogenetic protein tree. Only the nodes with ultrafast bootstrap (UFBoot) support values over 90% were labeled with black dots. The positive Sdo sequences that were checked by two conservative amino acid residues were labeled yellow in the tree. Three positive Sdo clades (including ETHE1, Sdo, and Blh) were labeled yellow; the numbers of positive Sdo sequences, non-Sdo sequences, and Sdo reference sequences were labeled accordingly. Other unannotated clades and non-Sdo clades (including metallo-beta-lactamase, GloB1, and GloB2) all contained non-Sdo sequences. Fig. S12. Relative abundance of GTDB-assigned MAG taxa at Guaymas Basin. Abundances are shown (A) for all taxa at the genus level, and (B) for the Archaea at the order level, using read coverage normalized to 100M reads per sample and expressed as a percentage of MAG reads per sample. Relative abundances were averaged for the two samples from the six-day thermocouple array (4561-380 and 4561-384).
Additional file 2: Table S1.
Sample metadata including location, year, research vessel, number of metagenome reads and accession numbers. Table S2. MAG genome properties, accession numbers and taxonomic classifications. Taxonomy was assigned using GTDB-Tk, and mis-classified MAGs were taxonomically re-assigned at the phylum level (Bacteria) and class level (Archaea) using curated archaeal and bacterial phylogenetic trees. Genome quality statistics are based on completion and contamination (high quality, >90% completion, <5% contamination; medium quality, ≥50% completion, ≤10% contamination). Average contamination was 4.02%. Table S3. Average amino acid identity (AAI) matrices for the (A) Bacteria and (B) Archaea. Matrices are grouped by GTDB taxonomy and include MAGs that could not be assigned to a known genus by GTDB-Tk. Details are provided which recently identified MAGs were from Brothers volcano hydrothermal deposits. Table S4. Relative abundance of GTDB taxa by site, based on read coverage of MAGs normalized to 100M reads per sample. MAG coverage for each site was summed and expressed as a percent. Table S5. METABOLIC-G results for JALSQH01 (3 MAGs) and JALWCF01 (13 MAGs). In the summary rows for JALSQH01 and JALWCF01, functions and modules are listed as “present” if identified in ≥50% of all MAGs, “partially present” if found in <50% of the MAGs, and “absent” if undetected in the MAGs. Table S6. Selected functional genes found in Patescibacteria MAGs, based on annotation with GhostKOALA. KEGG module numbers are shown in parentheses. Table S7. Functional genes identified in selected > 80%-completeness MAGs from the Chloroflexota. (A) Genes are marked as present (1; green highlight) or not detected (0) in individual MAGs. (B) The proportion of > 80%-completeness MAGs in six GTDB orders that encode functional genes is also shown, with proportions ≥50% highlighted in green. Table S8. Identification and distribution of functional genes in this study. (A) The HMMs, MEROPS peptidases, and CAZymes used to identify functional genes. Gene call numbers were calculated using the component (C) or accumulative (A) methods described in the methods. Genes requiring manual validation (M) are indicated. (B) Functional gene abundance, calculated as described in the methods. Table S9. Percentage of MAGs in phylogenetic clusters that encode core metabolic genes. Unless otherwise indicated, Archaea are shown at the class level, and Bacteria are shown at the phylum level. Genes were detected using METABOLIC, with additional validation steps for oxidative and reductive Dsr, Sdo, PmoA and McrA. Table S10. Comparative (A) relative abundance and (B) functional gene abundance for the Gammaproteobacteria and Campylobacteria, used to generate Fig. 10.
Additional file 3: Data S1.
Newick format archaeal concatenated protein phylogenetic tree, including both MAGs and GTDB reference genomes.
Additional file 4: DataS2.
Newick file ofbacterial concatenated protein phylogenetic tree including MAGs and GTDBreference genomes, generated using IQ-TREE.
Additional file 5: Data S3.
Concatenated protein phylogenetic tree of bacterial MAGs and GTDB reference genomes, generated with FastTree (Newick format).
Additional file 6:Data S4.
MAG-only bacterialconcatenated phylogenetic protein tree in Newick format.
Additional file 7: Data S5.
Concatenated protein phylogeny of archaeal MAGs in Newick format.
Additional file 8.
Supplementary Discussion.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Zhou, Z., St. John, E., Anantharaman, K. et al. Global patterns of diversity and metabolism of microbial communities in deep-sea hydrothermal vent deposits. Microbiome 10 , 241 (2022). https://doi.org/10.1186/s40168-022-01424-7
Download citation
Received : 05 August 2022
Accepted : 11 November 2022
Published : 27 December 2022
DOI : https://doi.org/10.1186/s40168-022-01424-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Thermophiles
- Deep-sea hydrothermal vents
- Metagenomics
ISSN: 2049-2618
- General enquiries: [email protected]
NTRS - NASA Technical Reports Server
Available downloads, related records.
- ARC Strategy
- Our Purpose
- Client Service Charter
- Corporate Reporting and Plans
- Feedback and Complaints
- Freedom of Information
- Work for us
- Corporate Policies
- Program Policies and Statements
- Find information
- Applying for a grant?
- Research Security
- Grant announcement kits
- Grants Dataset
- Selection outcome reports
- Applicant Feedback
- Grants Calendar
- Peer review
GrantConnect
Research grants services.
- The National Interest Test statement
- Manage your grant
- Previous rounds
- Data portal
- Terms and conditions
- Contact Officers
- Indigenous Studies
- ERA EI Review
- Consultation
- Explore University Partner
- Sponsorship arrangements/ collaborations
- Security Vulnerability Disclosure Policy
- Research Integrity Policy
- Codes and Guidelines
- Australian Research Integrity Committee
- Research Integrity in Australia - roles and responsibilities
- ARC Articles
- Articles from other sources
- Social media
- Publications
- Network messages
- Funding announcements (RMS)
Popular Searches
Below are some popular search terms you may find useful.
Data Portal

Expression Of Interest outcomes for Discovery Projects 2025

The Australian Research Council (ARC) has released outcomes of Expression of Interest applications for the Discovery Projects scheme (DP25).
All lead Chief Investigators will receive an email from RMS informing them of the outcome of their DP25 EOI application.
Successful and unsuccessful applicants will be able to access their scores in the feedback tab of their application in RMS.
Further information is available on the ARC website: https://www.arc.gov.au/funding-research/discovery-linkage/discovery-program/discovery-projects#key-documents
Full applications for shortlisted applicants open in RMS on Thursday 11 April 2024 and close at 5pm AEST on Tuesday 4 June 2024.
If you have any questions, please email [email protected] or phone +61 2 6287 6600.

COMMENTS
Checklist: Research results 0 / 7. I have completed my data collection and analyzed the results. I have included all results that are relevant to my research questions. I have concisely and objectively reported each result, including relevant descriptive statistics and inferential statistics. I have stated whether each hypothesis was supported ...
Begin with a clear statement of the principal findings. This will reinforce the main take-away for the reader and set up the rest of the discussion. Explain why the outcomes of your study are important to the reader. Discuss the implications of your findings realistically based on previous literature, highlighting both the strengths and ...
Tips to Write the Results Section. Direct the reader to the research data and explain the meaning of the data. Avoid using a repetitive sentence structure to explain a new set of data. Write and highlight important findings in your results. Use the same order as the subheadings of the methods section.
Research results refer to the findings and conclusions derived from a systematic investigation or study conducted to answer a specific question or hypothesis. These results are typically presented in a written report or paper and can include various forms of data such as numerical data, qualitative data, statistics, charts, graphs, and visual aids.
The discussion reviews the findings and puts them into the context of the overall research. It brings together all the sections that came before it and allows a reader to see the connections between each part of the research paper. In a discussion section, the author engages in three necessary steps: interpretation, analysis, and explanation.
The results section of a research paper tells the reader what you found, while the discussion section tells the reader what your findings mean. The results section should present the facts in an academic and unbiased manner, avoiding any attempt at analyzing or interpreting the data. Think of the results section as setting the stage for the ...
Papers usually end with a concluding section, often called the "Discussion.". The Discussion is your opportunity to evaluate and interpret the results of your study or paper, draw inferences and conclusions from it, and communicate its contributions to science and/or society. Use the present tense when writing the Discussion section.
A two-sample t test was used to test the hypothesis that higher social distance from environmental problems would reduce the intent to donate to environmental organisations, with donation intention (recorded as a score from 1 to 10) as the outcome variable and social distance (categorised as either a low or high level of social distance) as the predictor variable.Social distance was found to ...
II. The Content. The content of the discussion section of your paper most often includes:. Explanation of results: Comment on whether or not the results were expected for each set of findings; go into greater depth to explain findings that were unexpected or especially profound.If appropriate, note any unusual or unanticipated patterns or trends that emerged from your results and explain their ...
The discussion section provides an analysis and interpretation of the findings, compares them with previous studies, identifies limitations, and suggests future directions for research. This section combines information from the preceding parts of your paper into a coherent story. By this point, the reader already knows why you did your study ...
IMRaD Results Discussion. Results and Discussion Sections in Scientific Research Reports (IMRaD) After introducing the study and describing its methodology, an IMRaD* report presents and discusses the main findings of the study. In the results section, writers systematically report their findings, and in discussion, they interpret these findings.
This handout was created to accompany the Writing in the Sciences video series. The purpose of the Results is to prepare readers for the discussion section by presenting the data in manageable chunks, in an order that corresponds with the research questions or objectives. The purpose of the Discussion section is used to contain analysis and ...
Step 1: Consult the guidelines or instructions that the target journal or publisher provides authors and read research papers it has published, especially those with similar topics, methods, or results to your study. The guidelines will generally outline specific requirements for the results or findings section, and the published articles will ...
DISCUSSION. Evidence does not explain itself; the results must be presented and then explained. Typical stages in the discussion: summarizing the results, discussing whether results are expected or unexpected, comparing these results to previous work, interpreting and explaining the results (often by comparison to a theory or model), and hypothesizing about their generality.
Begin the Discussion section by restating your statement of the problem and briefly summarizing the major results. Do not simply repeat your findings. Rather, try to create a concise statement of the main results that directly answer the central research question that you stated in the Introduction section.
Discussion is mainly the section in a research paper that makes the readers understand the exact meaning of the results achieved in a study by exploring the significant points of the research, its ...
An example of research summary in discussion. 3.2. An example of result interpretation in discussion. 3.3. An example of literature comparison in discussion. 3.4. An example of research implications in discussion. 3.5. An example of limitations in discussion.
What not to include in your discussion section. There are a few common mistakes to avoid when writing the discussion section of your paper. Don't introduce new results: You should only discuss the data that you have already reported in your results section. Don't make inflated claims: Avoid overinterpretation and speculation that isn't directly supported by your data.
The discussion section of a research paper is where the author analyzes and explains the importance of the study's results. It presents the conclusions drawn from the study, compares them to previous research, and addresses any potential limitations or weaknesses.
The Results and Discussion sections can be written as separate sections (as shown in Fig. 2), but are often combined in a poster into one section called Results and Discussion. This is done in order to (1) save precious space on a poster for the many pieces of information that a scientist would like to tell their audience and (2) by combining the two sections, it becomes easier for the ...
This chapter 5 presents the results of the study. First, an outline of the informants included in the study and an overview of the statistical techniques employed in the data analyses are given ...
Similar results were observed in the high-risk subgroup (i.e., participants who had been vaccinated and had at least one risk factor for severe illness) and in the standard-risk subgroup (i.e ...
We conducted a noninferiority trial in which patients with clinically node-negative primary T1 to T3 breast cancer (tumor size, T1, ≤20 mm; T2, 21 to 50 mm; and T3, >50 mm in the largest ...
This paper describes a scoping review of China's academic resource databases, relevant official websites, news reports and public accounts spanning a period from the end of 2019 to the end of 2022, to investigate the challenges in scientific integrity and ethical soundness of research conducted during and immediately after the COVID-19 pandemic in China.
When deep-sea hydrothermal fluids mix with cold oxygenated fluids, minerals precipitate out of solution and form hydrothermal deposits. These actively venting deep-sea hydrothermal deposits support a rich diversity of thermophilic microorganisms which are involved in a range of carbon, sulfur, nitrogen, and hydrogen metabolisms. Global patterns of thermophilic microbial diversity in deep-sea ...
A conceptual architecture for retrieval of a sample of the surface of Venus is proposed. The mission concept incorporates a high-temperature aircraft to retrieve the sample from the surface and raise it into the upper atmosphere, a balloon-borne platform to produce fuel from the carbon dioxide atmosphere of Venus, and a launch vehicle to bring the sample into Venus orbit, where it is retrieved ...
A Feature Paper should be a substantial original Article that involves several techniques or approaches, provides an outlook for future research directions and describes possible research applications. Feature papers are submitted upon individual invitation or recommendation by the scientific editors and must receive positive feedback from the ...
The Australian Research Council (ARC) has released outcomes of Expression of Interest applications for the Discovery Projects scheme (DP25). All lead Chief Investigators will receive an email from RMS informing them of the outcome of their DP25 EOI application. Successful and unsuccessful applicants will be able to access their scores in the ...