Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery
Affiliations.
- 1 Structure-Based Drug Design Group, Organic Synthesis Department, Daiichi Sankyo RD Novare Co., Ltd, 1-16-13 Kita-Kasai, Edogawa-ku, Tokyo, 134-8630, Japan. [email protected].
- 2 Structure-Based Drug Design Group, Organic Synthesis Department, Daiichi Sankyo RD Novare Co., Ltd, 1-16-13 Kita-Kasai, Edogawa-ku, Tokyo, 134-8630, Japan.
- 3 Graduate School of Medical Life Science, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama, 230-0045, Japan.
- PMID: 32306215
- DOI: 10.1007/s10858-020-00314-0
Recently, there has been increasing interest in new modalities such as therapeutic antibodies and gene therapy at a number of pharmaceutical companies. Moreover, in small-molecule drug discovery at such companies, efforts have focused on hard-to-drug targets such as inhibiting protein-protein interactions. Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways, such as for the reliable detection of binding and providing three-dimensional structural information for structure-based drug design. The advantages of using NMR spectroscopy have been known for decades (Jahnke in J Biomol NMR 39:87-90, (2007); Gossert and Jahnke in Prog Nucl Magn Reson Spectrosc 97:82-125, (2016)). For tackling hard-to-drug targets and increasing the success in discovering drug molecules, in-depth analysis of drug-target protein interactions performed by biophysical methods will be more and more essential. Here, we review the advantages of NMR spectroscopy as a key technology of biophysical methods and also discuss issues such as using cutting-edge NMR spectrometers and increasing the demand of utilizing conformational dynamics information for promoting small-molecule drug discovery.
Keywords: Conformational analysis; Drug discovery; FBDD; Hit validation; SBDD.

Publication types
- Calorimetry, Differential Scanning
- Crystallography, X-Ray
- Drug Design
- Drug Discovery / methods*
- High-Throughput Screening Assays
- Molecular Docking Simulation
- Nuclear Magnetic Resonance, Biomolecular / methods*
- Protein Binding
- Quantitative Structure-Activity Relationship
- Small Molecule Libraries
- Share on Twitter
- Share on Facebook
- Share by email
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery
Mentioned by, x demographics, geographical breakdown, demographic breakdown, mendeley readers, attention score in context.
This page is provided by Altmetric .

- Structure Summary
- Annotations
- FASTA Sequence
- mmCIF Format
- mmCIF Format (Header)
- PDB Format (Header)
- PDBx/mmCIF Format
- PDBx/mmCIF Format (gz)
- BinaryCIF Format (gz)
- PDB Format (gz)
- PDBML/XML Format (gz)
- Structure Factors (CIF)
- Structure Factors (CIF - gz)
- Validation Full PDF
- Validation (XML - gz)
- Validation (CIF - gz)
- Biological Assembly 1 (CIF - gz)
- Biological Assembly 1 (PDB - gz)
- fo-fc Map (DSN6)
- 2fo-fc Map (DSN6)
- Map Coefficients (MTZ format)
Human PUF60 UHM domain (thioredoxin fusion) in complex with a small molecule binder
- PDB DOI: https://doi.org/10.2210/pdb6LUR/pdb
- Classification: SPLICING
- Organism(s): Escherichia coli K-12 , Homo sapiens
- Expression System: Escherichia coli
- Mutation(s): No
- Deposited: 2020-01-30 Released: 2020-04-29
- Deposition Author(s): Takahashi, M. , Hanzawa, H.
Experimental Data Snapshot
- Method: X-RAY DIFFRACTION
- Resolution: 2.00 Å
- R-Value Free: 0.253
- R-Value Work: 0.201
- R-Value Observed: 0.203
wwPDB Validation 3D Report Full Report
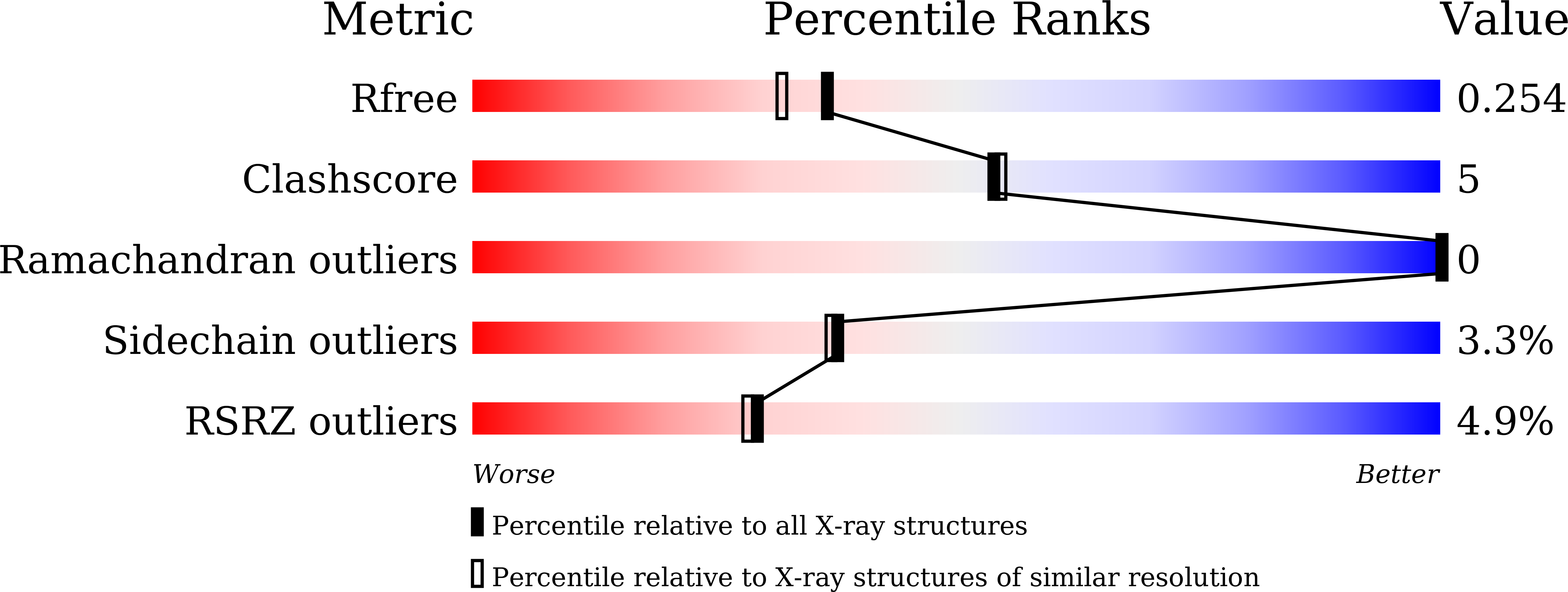
Ligand Structure Quality Assessment
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery.
(2020) J Biomol NMR 74 : 501-508
- PubMed : 32306215 Search on PubMed
- DOI: https://doi.org/10.1007/s10858-020-00314-0
- Primary Citation of Related Structures: 6LUR
Recently, there has been increasing interest in new modalities such as therapeutic antibodies and gene therapy at a number of pharmaceutical companies. Moreover, in small-molecule drug discovery at such companies, efforts have focused on hard-to-drug targets such as inhibiting protein-protein interactions. Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways, such as for the reliable detection of binding and providing three-dimensional structural information for structure-based drug design. The advantages of using NMR spectroscopy have been known for decades (Jahnke in J Biomol NMR 39:87-90, (2007); Gossert and Jahnke in Prog Nucl Magn Reson Spectrosc 97:82-125, (2016)). For tackling hard-to-drug targets and increasing the success in discovering drug molecules, in-depth analysis of drug-target protein interactions performed by biophysical methods will be more and more essential. Here, we review the advantages of NMR spectroscopy as a key technology of biophysical methods and also discuss issues such as using cutting-edge NMR spectrometers and increasing the demand of utilizing conformational dynamics information for promoting small-molecule drug discovery.
Structure-Based Drug Design Group, Organic Synthesis Department, Daiichi Sankyo RD Novare Co., Ltd, 1-16-13 Kita-Kasai, Edogawa-ku, Tokyo, 134-8630, Japan. [email protected].
Explore in 3D : Structure | Sequence Annotations | Electron Density | Validation Report | Ligand Interaction (EVU)
Biological assembly 1 assigned by authors and generated by PISA (software)
Macromolecule Content
- Total Structure Weight: 199.63 kDa
- Atom Count: 13,950
- Modelled Residue Count: 1,690
- Deposited Residue Count: 1,776
- Unique protein chains: 1
Experimental Data
- Space Group: P 2 1 2 1 2 1
Structure Validation
View Full Validation Report
Ligand Structure Quality Assessment
Deposition data.
- Released Date: 2020-04-29
Revision History (Full details and data files)
- Version 1.0: 2020-04-29 Type: Initial release
- Version 1.1: 2020-05-06 Changes: Database references
- Version 1.2: 2020-12-09 Changes: Database references
- Version 1.3: 2023-11-29 Changes: Data collection, Database references, Refinement description
- Publications
- Usage & Privacy
- Documentation
- Website FAQ
- Service Status
RCSB PDB ( citation ) is hosted by

RCSB PDB is a member of the

- RCSB Partners
- Nucleic Acid Knowledgebase
- wwPDB Partners
RCSB PDB Core Operations are funded by the U.S. National Science Foundation (DBI-2321666), the US Department of Energy (DE-SC0019749), and the National Cancer Institute , National Institute of Allergy and Infectious Diseases , and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM133198.
NMR as a Tool to Target Protein–Protein Interactions
- First Online: 01 January 2013
Cite this chapter

- Rebecca Del Conte 2 ,
- Daniela Lalli 2 &
- Paola Turano 2
786 Accesses
NMR spectroscopy plays a dual role in projects aimed at targeting protein–protein interactions (PPIs).While it has been extensively validated as an efficient technique for the initial screening and identification of weakly interacting fragments and for subsequently guiding their optimization into molecules with higher affinity and more favorable drug-like properties, it also represents an extremely powerful tool to monitor the formation of protein–protein complexes in solution and to obtain structural information on these adducts. It allows the identification of the protein interfaces and, in some cases, provides intermolecular distance and orientational restraints that lead to the definition of the relative arrangement of the two proteins. In particular, it constitutes the structural technique of choice for studying weak/ transient protein–protein interactions, which represent the natural targets for drug discovery projects addressing PPIs.
This is a preview of subscription content, log in via an institution to check access.
Access this chapter
- Available as EPUB and PDF
- Read on any device
- Instant download
- Own it forever
- Compact, lightweight edition
- Dispatched in 3 to 5 business days
- Free shipping worldwide - see info
- Durable hardcover edition
Tax calculation will be finalised at checkout
Purchases are for personal use only
Institutional subscriptions
Rowe AJ (2011) Ultra-weak reversible protein–protein interactions. Methods 54:157–166
Article CAS Google Scholar
Schreiber G, Haran G (2009) Zhou HX (2009) Fundamental aspects of protein–protein association kinetics. Chem Rev 109:839–860
Alsallaq R, Zhou HX (2008) Electrostatic rate enhancement and transient complex of protein–protein association. Proteins 71:320–335
Archakov AI, Govorun VM, Dubanov AV, Ivanov YD, Veselovsky AV, Lewi P, Janssen P (2003) Protein–protein interactions as a target for drugs in proteomics. Proteomics 3:380–391
Prudencio M, Ubbink M (2004) Transient complexes of redox proteins: structural and dynamic details from NMR studies. J Mol Recognit 17:524–539
Corzo J (2006) Time, the forgotten dimension of ligand binding teaching. Biochem Mol Biol Educ 34:413–416
Jensen MR, Ortega-Roldan JL, Salmon L, van Nuland N, Blackledge M (2011) Characterizing weak protein–protein complexes by NMR residual dipolar couplings. Eur Biophys J 40:1371–1381
Takeuchi K, Wagner G (2006) NMR studies of protein interactions. Curr Opin Struct Biol 16:109–117
Bonvin AM, Boelens R, Kaptein R (2005) NMR analysis of protein interactions. Curr Opin Chem Biol 9:501–508
Zuiderweg ER (2002) Mapping protein–protein interactions in solution by NMR spectroscopy. Biochemistry 41:1–7
Crowley PB, Ubbink M (2003) Close encounters of the transient kind: protein interactions in the photosynthetic redox chain investigated by NMR spectroscopy. Acc Chem Res 36:723–730
Meyer B, Peters T (2003) NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew Chem Int Ed 42:864–890
Zerbe O, Mannhold R, Kubinyi H, Folkers G (2003) BioNMR in drug research. Wiley-VCH, Zurich
Google Scholar
Vaynberg J, Qin J (2006) Weak protein–protein interactions as probed by NMR spectroscopy. Trends Biotechnol 24:22–27
Garrett DS, Seok YJ, Peterkofsky A, Clore GM, Gronenborn AM (1997) Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the escherichia coli phosphotransferase system. Biochemistry 36:4393–4398
Shuker SB, Hajduk PJ, Meadows RP, Fesik SW (1996) Discovering high-affinity ligands for proteins: SAR by NMR. Science 274:1531–1534
de Vries SJ, van Dijk M, Bonvin AM (2010) The HADDOCK web server for data-driven biomolecular docking. Nat Protoc 5:883–897
Article Google Scholar
Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein–protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125:1731–1737
Pervushin K, Riek R, Wider G, Wüthrich K (1997) Attenuated T 2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Riek R, Wider G, Pervushin K, Wüthrich K (1999) Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc Natl Acad Sci USA 96:4918–4923
Fiaux J, Bertelsen EB, Horwich AL, Wüthrich K (2002) NMR analysis of a 900 K GroEL GroES complex. Nature 418:207–211
Griswold IJ, Dahlquist FW (2002) Bigger is better: megadalton protein NMR in solution. Nat Struct Biol 9:567–568
Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wütrich K (2005) Direct NMR observation of a substrate protein bound to the chaperonin. GroEL. Proc Natl Acad Sci USA 102: 12748-12753
Caillet-Saguy C, Piccioli M, Turano P, Izadi-Pruneyre N, Delepierre M, Bertini I, Lecroisey A (2009) Mapping the Interaction between the Hemophore HasA and its outer membrane receptor has R using CRINEPT-TROSY NMR spectroscopy. J Am Chem Soc 131:1736–1744
Debye PJW (1929) Polar molecules. Dover Publications Inc
Ikura M, Bax A (1992) Isotope-filtered 2D NMR of protein-peptide complex: study of Skeletal muscle myosin light chain kinase. J Am Chem Soc 114:2433–2440
Banci L, Bertini I, Cefaro C, Cenacchi L, Ciofi-Baffoni S, Felli IC, Gallo A, Gonnelli L, Luchinat E, Sideris DP, Tokatlidis K (2010) Molecular chaperone function of Mia40 triggers consecutive induced folding steps of the substrate in mitochondrial protein import. Proc Natl Acad Sci USA 107:20190–20195
Prestegard JH, Bougault CM, Kishore AI (2004) Residual dipolar couplings in structure determination of biomolecules. Chem Rev 104:3519–3540
Capozzi F, Casadei F, Luchinat C (2006) EF-hand protein dynamics and evolution of calcium signal transduction: an NMR view. J Biol Inorg Chem 11:949–962
Bertini I, Calderone V, Cerofolini L, Fragai M, Geraldes CFGC, Hermann P, Luchinat C, Parigi G, Teixeira JMC (2012) The catalytic domain of MMP-1 studied through tagged lanthanides. FEBS Lett 586:557–567
Volkov AN, Ubbink M, Van Nuland NAJ (2010) Mapping the encounter state of a transient protein complex by PRE NMR spectroscopy. J Biomol NMR 48:225–236
Williamson MP, Marion D, Wuthrich K (1984) Secondary structure in the solution conformation of the proteinase inhibitor IIA from bull seminal plasma by nuclear magnetic resonance. J Mol Biol 173:341–359
O’Connell MR, Gamsjaeger R, Mackay JP (2009) The structural analysis of protein–protein interactions by NMR spectroscopy. Proteomics 9:5224–5232
Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu Y, Ng L, Wu C, Qin J (2005) Structure of an ultraweak protein–protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell 17:513–523
Wang JH, Meijers R, Xiong Y, Liu JH, Sakihama T, Zhang R, Joachimiak A, Reinherz EL (2001) Crystal structure of the human CD4N-terminal two-domain fragment complexed to a class II MHC molecule. Proc Natl Acad Sci USA 98:10799–10804
Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I (2003) Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell 113:621–630
Sundquist WI, Schubert HL, Kelly BN, Hill GC, Holton JM, Hill CP (2004) Ubiquitin recognition by the human TSG101 protein. Mol Cell 13:783–789
Ortega-Roldan JL, Jensen MR, Brutscher B, Azuaga AI, Blackledge M, van Nuland NA (2009) Accurate characterization of weak macromolecular interactions by titration of NMR residual dipolar couplings: application to the CD2AP SH3-C: ubiquitin complex. Nucleic Acids Res 37:e70
Banci L, Bertini I, Calderone V, Della Malva N, Felli IC, Neri S, Pavelkova A, Rosato A (2009) Copper(I)-mediated protein–protein interactions result from suboptimal interaction surfaces. Biochem J 422:37–42
Banci L, Bertini I, McGreevy KS, Rosato A (2010) Molecular recognition in copper trafficking. Nat Prod Rep 27:695–710
Banci L, Bertini I, Cantini F, Ciofi-Baffoni S (2010) Cellular copper distribution: a mechanistic systems biology approach. Cell Mol Life Sci 67:2563–2589
Guiles RD, Sarma S, DiGate RJ, Banville D, Basus VJ, Kuntz ID, Waskell L (1996) Pseudocontact shifts used in the restraint of the solution structures of electron transfer complexes. Nat Struct Biol 3:333–339
Ubbink M, Lian LY, Modi S, Evans PA, Bendall DS (1996) Analysis of the 1 H-NMR chemical shifts of Cu(I)−, Cu(II)- and Cd-substituted pea plastocyanin. Metal-dependent differences in the hydrogen-bond network around the copper site. Eur J Biochem 242:132–147
Hulsker R, Baranova MV, Bullerjahn GS, Ubbink M (2008) Dynamics in the transient complex of plastocyanin-cytochrome f from Prochlorothrix hollandica. J Am Chem Soc 130:1985–1991
Diaz-Moreno I, Diaz-Quintana A, De la Rosa MA, Ubbink M (2005) Structure of the complex between plastocyanin and cytochrome f from the cyanobacterium nostoc Sp. PCC 7119 as determined by paramagnetic NMR. J Biol Chem 280:18908–18915
Bashir Q, Scanu S, Ubbink M (2011) Dynamics in electron transfer protein complexes. FEBS J 278:1391–1400
Tang C, Iwahara J, Clore GM (2006) Visualization of transient encounter complexes in protein–protein association. Nature 444:383–386
Volkov AN, Worrall JAR, Holtzmann E, Ubbink M (2006) Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci USA 103:18945–18950
Xu X, Reinle W, Hannemann F, Konarev PV, Svergun DI, Bernhardt R, Ubbink M (2008) Dynamics in a pure encounter complex of two proteins studied by solution scattering and paramagnetic NMR spectroscopy. J Am Chem Soc 130:6395–6403
Liang ZX, Nocek JM, Huang K, Hayes RT, Kurnikov IV, Beratan DN, Hoffman BM (2002) Dynamic docking and electron transfer between Zn-myoglobin and cytochrome b(5). J Am Chem Soc 124:6849–6859
Volkov AN, Ferrari D, Worrall JA, Bonvin AM, Ubbink M (2005) The orientations of cytochrome c in the highly dynamic complex with cytochrome b5 visualized by NMR and docking using HADDOCK. Protein Sci 14:799–811
Liang ZX, Kurnikov IV, Nocek JM, Mauk AG, Beratan DN, Hoffman BM (2004) Dynamic docking and electron-transfer between cytochrome b5 and a suite of myoglobin surface-charge mutants. Introduction of a functional-docking algorithm for protein–protein complexes. J Am Chem Soc 126:2785–2798
Worrall JA, Liu A, Crowley PB, Nocek JM, Hoffman BM, Ubbink M (2002) Myoglobin and cytochrome b5: a nuclear magnetic resonance study of a highly dynamic protein complex. Biochemistry 41:11721–11730
Worrall JA, Reinle W, Bernhardt R, Ubbink M (2003) Transient protein interactions studied by NMR spectroscopy: the case of cytochrome C and adrenodoxin. Biochemistry 42:7068–7076
Hoffman BM, Celis LM, Cull DA, Patel AD, Seifert JL, Wheeler KE, Wang J, Yao J, Kurnikov IV, Nocek JM (2005) Differential influence of dynamic processes on forward and reverse electron transfer across a protein–protein interface. Proc Natl Acad Sci USA 102:3564–3569
Ubbink M, Bendall DS (1997) Complex of plastocyanin and cytochrome c characterized by NMR chemical shift analysis. Biochemistry 36:6326–6335
Vlasie MD, Fernández-Busnadiego R, Prudêncio M, Ubbink M (2008) Conformation of pseudoazurin in the 152 kDa electron transfer complex with nitrite reductase determined by paramagnetic NMR. J Mol Biol 375:1405–1415
Ubbink M, Ejdebaeck M, Karlsson BG, Bendall DS (1998) The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics. Structure 6:323–335
Bashir Q, Volkov AN, Ullmann GM, Ubbink M (2010) Visualization of the encounter ensemble of the transient electron transfer complex of cytochrome c and cytochrome c peroxidase. J Am Chem Soc 132:241–247
Fawzi NL, Doucleff M, Suh JY, Clore GM (2010) Mechanistic details of a protein–protein association pathway revealed by paramagnetic relaxation enhancement titration measurements. Proc Natl Acad Sci USA 107:1379–1384
Villareal VA, Spirig T, Robson SA, Liu M, Lei B, Clubb RT (2011) Transient weak protein–protein complexes transfer heme across the cell wall of Staphylococcus aureus. J Am Chem Soc 133:14176–14179
Nooren IMA, Thornton JM (2003) Structural characterisation and functional significance of transient protein–protein interactions. J Mol Biol 325:991–1018
Veselovsky AV, Archakov AI (2007) Inhibitors of protein–protein interactions as potential drugs. Curr Comput: Aided Drug Des 3:51–58
Arkin MR, Wells JA (2004) Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat Rev Drug Discov 3:301–317
Clackson T, Wells JA (1995) A hot spot of binding energy in a hormone-receptor interface. Science 267:383–386
Bogan AA, Thorn KS (1998) Anatomy of hot spots in protein interfaces. J Mol Biol 280:1–9
Agamennone M, Cesari L, Lalli D, Turlizzi E, Del Conte R, Turano P, Mangani S, Padova A (2010) Fragmenting the S100B–p53 interaction—Combined virtual/biophysical screening approaches to identify ligands. Chem Med Chem 5:428–435
CAS Google Scholar
Rustandi RR, Baldisseri DM, Weber DJ (2000) Structure of the negative regulatory domain of p53 bound to S100B (betabeta). Nat Struct Biol 7:570–574
Inman KG, Yang R, Rustandi RR, Miller KE, Baldisseri DM, Weber DJ (2002) Solution NMR structure of S100B bound to the high-affinity target peptide TRTK-12. J Mol Biol 324:1003–1014
Ivanenkov VV, Jamieson GA Jr, Gruenstein E, Dimlich RV (1995) Characterization of S-100b binding epitopes. Identification of a novel target, the actin capping protein, CapZ. J Biol Chem 270:14651–14658
Gaffen SL (2001) Signaling domains of the interleukin 2 receptor. Cytokine 14:63–77
Teague SJ (2003) Implications of protein flexibility for drug discovery. Nat Rev Drug Discov 2:527–541
Zhong S, Macias AT, Mackerell AD Jr (2007) Computational identification of inhibitors of protein–protein interactions. Curr Top Med Chem 7:63–82
Gonzalez-Ruiz D, Gohlke H (2006) Targeting protein–protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem 13:2607–2625
DeLano WL (2002) Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol 12:14–20
Kay LE (1998) Protein dynamics from NMR. Nat Struct Biol 5:513–517
Ishima R, Torchia DA (2000) Protein dynamics from NMR. Nat Struct Biol 7:740–743
Pellecchia M, Bertini I, Cowburn D, Dalvit C, Giralt E, Jahnke W, James TL, Homans SW, Kessler H, Luchinat C, Meyer B, Oschkinat H, Peng J, Schwalbe H, Siegal G (2008) Perspectives on NMR in drug discovery: a technique comes of age. Nat Rev Drug Discov 7:738–745
Stark JL, Powers R (2011) Application of NMR and molecular docking in structure-based drug discovery. Top Curr Chem. doi: 10.1007/128_2011_213
Fesik SW, Zuiderweg ER, Olejniczak ET, Gampe RT Jr (1990) NMR methods for determining the structures of enzyme/inhibitor complexes as an aid in drug design. Biochem Pharmacol 40:161–167
Meinecke R, Meyer B (2001) Determination of the binding specificity of an integral membrane protein by saturation transfer difference NMR: RGD peptide ligands binding to integrin alpha IIb beta 3 . J Med Chem 44:3059-3065
Wang YS, Liu D, Wyss DF (2004) Competition STD NMR for the detection of high-affinity ligands and NMR-based screening. Magn Reson Chem 42:485–489
Dalvit C, Fogliatto G, Stewart A, Veronesi M, Stockman BJ (2001) Water LOGSY as a method for primary NMR screening: practical aspects and range of applicability. J Biomol NMR 21:349–359
Wu D, Chen A, Johnson CS (1995) An improved diffusion-ordered spectroscopy experiment incorporating bipolar-gradient pulses. J Magn Reson A 115:260–264
Dalvit C, Fasolini M, Flocco M, Knapp S, Pevarello P, Veronesi M (2002) NMR-based screening with competition water-ligand observed via gradient spectroscopy experiments: detection of high-affinity ligands. J Med Chem 45:2610–2614
Assfalg M, Bertini I, Del Conte R, Giachetti A, Turano P (2007) Cytochrome c and organic molecules: the solution structure of the para-aminophenol adduct. Biochemistry 46:6232–6238
Bertini I, Calderone V, Cosenza M, Fragai M, Lee Y-M, Luchinat C, Mangani S, Terni B, Turano P (2005) Conformational variability of MMPs: beyond a single 3D structure. Proc Natl Acad Sci USA 102:5334–5339
Isaksson J, Nystroem S, Derbishire W, Wallberg H, Agback T, Kovacs H, Bertini I, Giachetti A, Luchinat C (2009) Does a fast nuclear magnetic resonance spectroscopy- and X-ray crystallography hybrid approach provide reliable structural information of ligand-protein complexes? A case study of metalloproteinases. J Med Chem 52:1712–1722
Constantine KL, Davis ME, Metzler WJ, Mueller L, Claus BL (2006) Protein-ligand NOE matching: a high-throughput method for binding pose evaluation that does not require protein NMR resonance assignments. J Am Chem Soc 128:7252–7263
Adams JM, Cory S (1998) The Bcl-2 protein family: arbiters of cell survival. Science 281:1322–1326
Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW (1997) Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275:983–986
Petros AM, Nettesheim DG, Wang Y, Olejniczak ET, Meadows RP, Mack J, Swift K, Matayoshi ED, Zhang H, Thompson CB, Fesik SW (2000) Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci 9:2528–2534
Petros AM, Dinges J, Augeri DJ, Baumeister SA, Betebenner DA, Bures MG, Elmore SW, Hajduk PJ, Joseph MK, Landis SK, Nettesheim DG, Rosenberg SH, Shen W, Thomas S, Wang X, Zanze I, Zhang H, Fesik SW (2006) Discovery of a potent inhibitor of the antiapoptotic protein Bcl-xL from NMR and parallel synthesis. J Med Chem 49:656–663
Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng S, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435:677–681
Petros AM, Huth JR, Oost T, Park CM, Ding H, Wang X, Zhang H, Nimmer P, Mendoza R, Sun C, Mack J, Walter K, Dorwin S, Gramling E, Ladror U, Rosenberg SH, Elmore SW, Fesik SW, Hajduk PJ (2010) Discovery of a potent and selective Bcl-2 inhibitor using SAR by NMR. Bioorg Med Chem Lett 20:6587–6591
Bertini I, Chevance S, Del Conte R, Lalli D, Turano P (2011) The anti-apoptotic Bcl-xL protein, a new piece in the puzzle of cytochrome c interactome. PLoS ONE 6:e18329
Arendt Y, Bhaumik A, Del Conte R, Luchinat C, Mori M, Porcu M (2007) Fragment docking to S100 proteins reveals a wide diversity of weak interaction sites. Chem Med Chem 2:1648–1654
Download references
Author information
Authors and affiliations.
Magnetic Resonance Center (CERM), University of Florence, Via Luigi Sacconi 6, 50019, Florence, Sesto Fiorentino, Italy
Rebecca Del Conte, Daniela Lalli & Paola Turano
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Paola Turano .
Editor information
Editors and affiliations.
, Department of Biotechnology, Chemistry a, University of Siena, Siena, Siena, I-53100, Italy
Stefano Mangani
Rights and permissions
Reprints and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg

About this chapter
Del Conte, R., Lalli, D., Turano, P. (2013). NMR as a Tool to Target Protein–Protein Interactions. In: Mangani, S. (eds) Disruption of Protein-Protein Interfaces. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-37999-4_4
Download citation
DOI : https://doi.org/10.1007/978-3-642-37999-4_4
Published : 28 June 2013
Publisher Name : Springer, Berlin, Heidelberg
Print ISBN : 978-3-642-37998-7
Online ISBN : 978-3-642-37999-4
eBook Packages : Chemistry and Materials Science Chemistry and Material Science (R0)
Share this chapter
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Publish with us
Policies and ethics
- Find a journal
- Track your research
Europe PMC requires Javascript to function effectively.
Either your web browser doesn't support Javascript or it is currently turned off. In the latter case, please turn on Javascript support in your web browser and reload this page.
Search life-sciences literature (44,031,988 articles, preprints and more)
- Available from publisher site using DOI. A subscription may be required. Full text
- Citations & impact
- Similar Articles
NMR spectroscopy: the swiss army knife of drug discovery.
Author information, affiliations.
- Farley KA 1
- Kormos BL 2
- Withka JM 2
ORCIDs linked to this article
- Farley KA | 0000-0001-8935-6852
- Horst R | 0000-0002-2684-0310
Journal of Biomolecular NMR , 02 Jul 2020 , 74(10-11): 509-519 https://doi.org/10.1007/s10858-020-00330-0 PMID: 32617727
Abstract
Full text links .
Read article at publisher's site: https://doi.org/10.1007/s10858-020-00330-0
Citations & impact
Impact metrics, alternative metrics.

Article citations
11 b nmr of the morphological evolution of traditional chinese medicine borax..
Li Q , Yang Y , Wang Q , Han X , Zhu J , Zhang N , Wang Q , Li K , Gong P , Chen F
Molecules , 29(1):251, 03 Jan 2024
Cited by: 0 articles | PMID: 38202834 | PMCID: PMC10780283
Endogenous modulators of neurotrophin signaling: Landscape of the transient ATP-NGF interactions.
Paoletti F , Merzel F , Cassetta A , Ogris I , Covaceuszach S , Grdadolnik J , Lamba D , Golič Grdadolnik S
Comput Struct Biotechnol J , 19:2938-2949, 07 May 2021
Cited by: 2 articles | PMID: 34136093 | PMCID: PMC8164016
Similar Articles
To arrive at the top five similar articles we use a word-weighted algorithm to compare words from the Title and Abstract of each citation.
Fragment-based drug discovery using NMR spectroscopy.
Harner MJ , Frank AO , Fesik SW
J Biomol NMR , 56(2):65-75, 18 May 2013
Cited by: 86 articles | PMID: 23686385 | PMCID: PMC3699969
Review Free full text in Europe PMC
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery.
Hanzawa H , Shimada T , Takahashi M , Takahashi H
J Biomol NMR , 74(10-11):501-508, 18 Apr 2020
Cited by: 3 articles | PMID: 32306215
Advancing fragment binders to lead-like compounds using ligand and protein-based NMR spectroscopy.
Methods Enzymol , 493:469-485, 01 Jan 2011
Cited by: 3 articles | PMID: 21371602
Counting on Fragment Based Drug Design Approach for Drug Discovery.
Kashyap A , Singh PK , Silakari O
Curr Top Med Chem , 18(27):2284-2293, 01 Jan 2018
Cited by: 4 articles | PMID: 30499406
Target immobilization as a strategy for NMR-based fragment screening: comparison of TINS, STD, and SPR for fragment hit identification.
Kobayashi M , Retra K , Figaroa F , Hollander JG , Ab E , Heetebrij RJ , Irth H , Siegal G
J Biomol Screen , 15(8):978-989, 01 Sep 2010
Cited by: 15 articles | PMID: 20817886
Europe PMC is part of the ELIXIR infrastructure

IMAGES
VIDEO
COMMENTS
Moreover, in small-molecule drug discovery at such companies, efforts have focused on hard-to-drug targets such as inhibiting protein-protein interactions. Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways, such as for the reliable detection of binding and providing three-dimensional structural information for ...
Recently, there has been increasing interest in new modalities such as therapeutic antibodies and gene therapy at a number of pharmaceutical companies. Moreover, in small-molecule drug discovery at such companies, efforts have focused on hard-to-drug targets such as inhibiting protein-protein interactions. Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways ...
The advantages of N MR spectroscopy as a key technology of biophysical methods are reviewed and issues such as using cutting-edge NMR spectrometers and increasing the demand of utilizing conformational dynamics information for promoting small-molecule drug discovery are discussed. Recently, there has been increasing interest in new modalities such as therapeutic antibodies and gene therapy at ...
Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways, such as for the reliable detection of binding and providing three-dimensional structural information for ...
This website requires cookies, and the limited processing of your personal data in order to function. By using the site you are agreeing to this as outlined in our privacy notice and cookie policy.
NMR spectroscopy is one of those technologies that has persisted for many decades, and continues to play an important and sometimes essential role, especially when dealing with difficult targets. The role of NMR in target-based drug discovery is manifold. It is an important technique for fragment-based lead discovery (FBLD), in particular for ...
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery. Hiroyuki Hanzawa; Takashi Shimada; Hideo Takahashi; Perspective 18 April 2020 Pages: 501 - 508 NMR spectroscopy: the swiss army knife of drug discovery. Reto Horst; Kathleen A. Farley; Jane M. Withka ...
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery. Journal of Biomolecular NMR 2020, 74 ... Protein—ligand structure determination with the NMR molecular replacement tool, NMR2. Journal of Biomolecular NMR 2020, 74 (10-11 ... Perspectives on NMR in drug discovery: a technique comes of age. Nature ...
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery Published in: Journal of Biomolecular NMR, April 2020 DOI: 10.1007/s10858-020-00314-0: Authors: Hiroyuki Hanzawa, Takashi Shimada, Mizuki Takahashi, Hideo Takahashi View on publisher site Alert me about new mentions.
Gossert AD, Jahnke W (2016) NMR in drug discovery: A practical guide to identification and validation of ligands interacting with biological macromolecules. Prog Nucl Magn Reson Spectrosc 97:82-125
Moreover, in small-molecule drug discovery at such companies, efforts have focused on hard-to-drug targets such as inhibiting protein-protein interactions. Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways, such as for the reliable detection of binding and providing three-dimensional structural information for ...
The study of small molecule-target interactions is central to small-molecule drug discovery. Biomolecular nuclear magnetic resonance (NMR) provides a wide array of approaches to study such interactions: ranging from functional readouts to biophysical measurements to atomic-level structural models.
Moreover, in small-molecule drug discovery at such companies, efforts have focused on hard-to-drug targets such as inhibiting protein-protein interactions. Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways, such as for the reliable detection of binding and providing three-dimensional structural information for ...
Pseudocontact Shifts in Biomolecular NMR Spectroscopy. Chemical Reviews 2022, 122 (10) ... Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery. Journal of Biomolecular NMR 2020, 74 (10-11) ...
These advances are allowing NMR to help solve important problems in the field of drug discovery. Their impact is widespread. NMR spectroscopy is now being used to determine protein structures, to monitor ligand-receptor binding, to study diffusion, to analyze mixtures using LC-NMR, to analyze solid-phase synthesis resins and to determine the ...
The roles of solution NMR spectroscopy in drug discovery are summarized, with some methods that are used in identifying fragments, understanding the mechanism of action for a ligand, and monitoring the conformational changes of a target induced by ligand binding. ... Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug ...
Revisiting a challenging p53 binding site: a diversity-optimized HEFLib reveals diverse binding modes in T-p53C-Y220C. ... Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery. Journal of Biomolecular NMR 2020, 74 ... the swiss army knife of drug discovery. Journal of Biomolecular NMR 2020, 74 (10-11) , 509-519 ...
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery. H. Hanzawa T. Shimada Mizuki Takahashi Hideo Takahashi. Chemistry, Medicine. ... as using cutting-edge NMR spectrometers and increasing the demand of utilizing conformational dynamics information for promoting small-molecule drug discovery are discussed.
Since the initial discovery of the magnetic resonance phenomenon in physics, NMR spectroscopy has evolved over the last 70 years, via widespread applications as an analytical tool in chemistry, into a versatile methodology that can address biological questions. This Thematic Issue on Biomolecular NMR Spectroscopy aims to bring recent advances ...
Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery ... This case history has been selected to supply the reader with an example of a successful application of NMR in drug discovery: here small organic molecules that bind to proximal subsites of a protein are identified, optimized and linked together to produce ...
Nuclear magnetic resonance (NMR) spectroscopy has evolved into a powerful tool within drug discovery over the last two decades. While traditionally being used by medicinal chemists for small molecule structure elucidation, it can also be a valuable tool for the identification of small molecules that bind to drug targets, for the characterization of target-ligand interactions and for hit-to ...
This perspectives article suggests key areas of impact for NMR, and a model of integrating NMR with other technologies to realize synergies and maximize their value for drug discovery. The versatility of NMR and its broad applicability to several stages in the drug discovery process is well known and generally considered one of the major strengths of NMR (Pellecchia et al., Nature Rev Drug ...
NMR is also an important tool in drug design and development. A number of successful drugs, available in the market or currently in clinical trials, originated from fragment-based drug design ...